QEI (Q-Noisy Expected Improvement) Demo for Blog
from qmcpy import *
import numpy as np
from scipy.linalg import solve_triangular, cho_solve, cho_factor
from scipy.stats import norm
import matplotlib.pyplot as pyplot
%matplotlib inline
lw = 3
ms = 8
Problem setup
Here is the current data (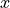 and
and  values with noise)
from which we want to build a GP and run a Bayesian optimization.
values with noise)
from which we want to build a GP and run a Bayesian optimization.
def yf(x):
return np.cos(10 * x) * np.exp(.2 * x) + np.exp(-5 * (x - .4) ** 2)
xplt = np.linspace(0, 1, 300)
yplt = yf(xplt)
x = np.array([.1, .2, .4, .7, .9])
y = yf(x)
v = np.array([.001, .05, .01, .1, .4])
pyplot.plot(xplt, yplt, linewidth=lw)
pyplot.plot(x, y, 'o', markersize=ms, color='orange')
pyplot.errorbar(x, y, yerr=2 * np.sqrt(v), marker='', linestyle='', color='orange', linewidth=lw)
pyplot.title('Sample data with noise');

Computation of the qEI quantity using qmcpy
One quantity which can appear often during BO is a computation involving
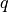 “next points” to sample in a BO process; in the standard
formulation this quantity might involve just
“next points” to sample in a BO process; in the standard
formulation this quantity might involve just 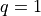 , but
, but
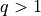 is also of interest for batched evaluation in parallel.
is also of interest for batched evaluation in parallel.
This quantity is defined as
![\mathrm{EI}_q(x_1, \ldots, x_q;\mathbf{y}, \mathcal{X}, \boldsymbol{\epsilon}) = \int_{\mathbb{R}^q} \max_{1\leq i\leq q}\left[{(y_i - y^*)_+}\right]\, p_{Y_{x_1,\ldots, x_q}|\mathbf{y}, \mathcal{X}, \boldsymbol{\epsilon}}(y_1, \ldots, y_q)\; \text{d}y_1\cdots\text{d}y_q](../_images/math/cf5cb8cf931987037b3d4ce8bd2c54c2a7758efa.png)
The example we are considering here is with 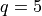 but this
quantity could be made larger. Each of these QEI computations (done in a
vectorized fashion in production) would be needed in an optimization
loop (likely powered by CMAES or some other high dimensional non-convex
optimization tool). This optimization problem would take place in a
but this
quantity could be made larger. Each of these QEI computations (done in a
vectorized fashion in production) would be needed in an optimization
loop (likely powered by CMAES or some other high dimensional non-convex
optimization tool). This optimization problem would take place in a
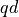 dimensional space, which is one aspect which usually prevents
dimensional space, which is one aspect which usually prevents
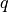 from being too large.
from being too large.
Note that some of this will look much more confusing in 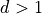 , but
it is written here in a simplified version.
, but
it is written here in a simplified version.
GP model definition (kernel information) and qEI definition
shape_parameter = 4.1
process_variance = .9
fudge_factor = 1e-10
def gaussian_kernel(x, z):
return process_variance * np.exp(-shape_parameter ** 2 * (x[:, None] - z[None, :]) ** 2)
def gp_posterior_params(x_to_draw):
n = len(x_to_draw)
kernel_prior_data = gaussian_kernel(x, x)
kernel_cross_matrix = gaussian_kernel(x_to_draw, x)
kernel_prior_plot = gaussian_kernel(x_to_draw, x_to_draw)
prior_cholesky = np.linalg.cholesky(kernel_prior_data + np.diag(v))
partial_cardinal_functions = solve_triangular(prior_cholesky, kernel_cross_matrix.T, lower=True)
posterior_covariance = kernel_prior_plot - np.dot(partial_cardinal_functions.T, partial_cardinal_functions) + fudge_factor * np.eye(n)
full_cardinal_functions = solve_triangular(prior_cholesky.T, partial_cardinal_functions, lower=False)
posterior_mean = np.dot(full_cardinal_functions.T, y)
return posterior_mean,posterior_covariance
def gp_posterior_draws(x_to_draw, mc_strat, num_posterior_draws, posterior_mean, posterior_covariance):
q = len(x_to_draw)
if mc_strat == 'iid':
dd = IIDStdUniform(q)
elif mc_strat == 'lattice':
dd = Lattice(q)
elif mc_strat == 'sobol':
dd = Sobol(q)
g = Gaussian(dd,posterior_mean,posterior_covariance)
posterior_draws = g.gen_samples(num_posterior_draws)
return posterior_draws
def compute_qei(posterior_draws):
y_gp = np.fmax(np.max(posterior_draws.T - max(y), axis=0), 0)
return y_gp
Demonstrate the concept of qEI on 2 points
num_posterior_draws = 2 ** 7
Np = (25, 24)
X, Y = np.meshgrid(np.linspace(0, 1, Np[1]), np.linspace(0, 1, Np[0]))
xp = np.array([X.reshape(-1), Y.reshape(-1)]).T
mu_post,sigma_cov = gp_posterior_params(xplt)
y_draws = gp_posterior_draws(xplt, 'lattice', num_posterior_draws,mu_post,sigma_cov).T
qei_vals = np.empty(len(xp))
for k, next_x in enumerate(xp):
mu_post,sigma_cov = gp_posterior_params(next_x)
gp_draws = gp_posterior_draws(next_x, 'sobol', num_posterior_draws,mu_post,sigma_cov)
qei_vals[k] = compute_qei(gp_draws).mean()
Z = qei_vals.reshape(Np)
fig, axes = pyplot.subplots(1, 3, figsize=(14, 4))
ax = axes[0]
ax.plot(xplt, yplt, linewidth=lw)
ax.plot(x, y, 'o', markersize=ms, color='orange')
ax.errorbar(x, y, yerr=2 * np.sqrt(v), marker='', linestyle='', color='orange', linewidth=lw)
ax.set_title('Sample data with noise')
ax.set_ylim((-2.3, 2.6))
ax = axes[1]
ax.plot(xplt, y_draws, linewidth=lw, color='b', alpha=.05)
ax.plot(x, y, 'o', markersize=ms, color='orange')
ax.errorbar(x, y, yerr=2 * np.sqrt(v), marker='', linestyle='', color='orange', linewidth=lw)
ax.set_title(f'{num_posterior_draws} GP posterior draws')
ax.set_ylim((-2.3, 2.6))
ax = axes[2]
h = ax.contourf(X, Y, Z)
ax.set_xlabel('First next point')
ax.set_ylabel('Second next point')
ax.set_title('qEI for q=2 next points')
cax = fig.colorbar(h, ax=ax)
cax.set_label('qEI')
fig.tight_layout()

Choose some set of next points against which to test the computation
Here, we consider 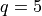 , which is much more costly to compute than
the
, which is much more costly to compute than
the  demonstration above.
demonstration above.
Note This will take some time to run. Use fewer num_repeats to
reduce the cost.
# parameters
next_x = np.array([0.158, 0.416, 0.718, 0.935, 0.465])
num_posterior_draws_to_test = 2 ** np.arange(4, 20)
d = len(next_x)
mu_post,sigma_cov = gp_posterior_params(next_x)
# get reference answer with qmcpy
integrand = CustomFun(
true_measure = Gaussian(Sobol(d),mu_post,sigma_cov),
g = compute_qei)
stopping_criterion = CubQMCSobolG(integrand, abs_tol=5e-7)
reference_answer,data = stopping_criterion.integrate()
print(data)
Solution: 0.0244
CustomFun (Integrand Object)
Sobol (DiscreteDistribution Object)
d 5
randomize 1
graycode 0
seed [68566 92413 32829 13022 69017]
mimics StdUniform
dim0 0
Gaussian (TrueMeasure Object)
mean [ 0.807 0.371 1.204 -0.455 0.67 ]
covariance [[ 1.845e-02 -2.039e-03 1.150e-04 1.219e-04 -5.985e-03]
[-2.039e-03 1.355e-02 6.999e-04 -1.967e-03 2.302e-02]
[ 1.150e-04 6.999e-04 8.871e-02 2.043e-02 5.757e-03]
[ 1.219e-04 -1.967e-03 2.043e-02 2.995e-01 -9.482e-03]
[-5.985e-03 2.302e-02 5.757e-03 -9.482e-03 6.296e-02]]
decomp_type pca
CubQMCSobolG (StoppingCriterion Object)
abs_tol 5.00e-07
rel_tol 0
n_init 2^(10)
n_max 2^(35)
LDTransformData (AccumulateData Object)
n_total 2^(22)
solution 0.024
error_bound 4.15e-07
time_integrate 4.426
# generate data
num_posterior_draws_to_test = 2 ** np.arange(4, 20)
vals = {}
num_repeats = 50
mc_strats = ('iid', 'lattice', 'sobol')
for mc_strat in mc_strats:
vals[mc_strat] = []
for num_posterior_draws in num_posterior_draws_to_test:
all_estimates = []
for _ in range(num_repeats):
y_draws = gp_posterior_draws(next_x, mc_strat, num_posterior_draws,mu_post,sigma_cov)
all_estimates.append(compute_qei(y_draws).mean())
vals[mc_strat].append(all_estimates)
vals[mc_strat] = np.array(vals[mc_strat])
fig, ax = pyplot.subplots(1, 1, figsize=(6, 4))
colors = ('#F5811F', '#A23D97', '#00B253')
alpha = .3
for (name, results), color in zip(vals.items(), colors):
bot = np.percentile(abs(results - reference_answer), 25, axis=1)
med = np.percentile(abs(results - reference_answer), 50, axis=1)
top = np.percentile(abs(results - reference_answer), 75, axis=1)
ax.loglog(num_posterior_draws_to_test, med, label=name, color=color)
ax.fill_between(num_posterior_draws_to_test, bot, top, color=color, alpha=alpha)
ax.loglog(num_posterior_draws_to_test, .1 * num_posterior_draws_to_test ** -.5, '--k', label='$O(N^{-1/2})$')
ax.loglog(num_posterior_draws_to_test, .25 * num_posterior_draws_to_test ** -1.0, '-.k', label='$O(N^{-1})$')
ax.set_xlabel('N - number of points')
ax.set_ylabel('Accuracy')
ax.legend(loc='lower left')
ax.set_title(f'Statistics from {num_repeats} runs');
# plt.savefig('qei_convergence.png');

# parameters
names = ['IID','Lattice','Sobol']
epsilons = [
[2e-4, 5e-4, 1e-3, 2e-3, 5e-3, 1e-2], # iid nodes
[5e-6, 1e-5, 2e-5, 5e-5, 1e-4, 2e-4, 5e-4, 1e-3, 2e-3, 5e-3, 1e-2], # lattice
[5e-6, 1e-5, 2e-5, 5e-5, 1e-4, 2e-4, 5e-4, 1e-3, 2e-3, 5e-3, 1e-2]] # sobol
trials = 25
# initialize time data
times = {names[j]:np.zeros((len(epsilons[j]),trials),dtype=float) for j in range(len(names))}
n_needed = {names[j]:np.zeros((len(epsilons[j]),trials),dtype=float) for j in range(len(names))}
# run tests
for t in range(trials):
print(t)
for j in range(len(names)):
for i in range(len(epsilons[j])):
if j == 0:
sc = CubMCG(CustomFun(Gaussian(IIDStdUniform(d),mu_post,sigma_cov),compute_qei),abs_tol=epsilons[j][i],rel_tol=0)
elif j == 1:
sc = CubQMCLatticeG(CustomFun(Gaussian(Lattice(d),mu_post,sigma_cov),compute_qei),abs_tol=epsilons[j][i],rel_tol=0)
else:
sc = CubQMCSobolG(CustomFun(Gaussian(Sobol(d),mu_post,sigma_cov),compute_qei),abs_tol=epsilons[j][i],rel_tol=0)
solution,data = sc.integrate()
times[names[j]][i,t] = data.time_integrate
n_needed[names[j]][i,t] = data.n_total
0
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
fig,axs = pyplot.subplots(1, 3, figsize=(22, 6))
colors = ('#245EAB', '#A23D97', '#00B253')
light_colors = ('#A3DDFF', '#FFBCFF', '#4DFFA0')
alpha = .3
def plot_fills(eps,data,name,color,light_color):
bot = np.percentile(data, 5, axis=1)
med = np.percentile(data, 50, axis=1)
top = np.percentile(data, 95, axis=1)
ax.loglog(eps, med, label=name, color=color)
ax.fill_between(eps, bot, top, color=light_color)
return med
for i,(nt_data,label) in enumerate(zip([times,n_needed],['time','n'])):
ax = axs[i+1]
# iid plot
eps_iid = np.array(epsilons[0])
data = nt_data['IID']
med_iid = plot_fills(eps_iid,data,'IID',colors[0],light_colors[0])
# lattice plot
eps = np.array(epsilons[1])
data = nt_data['Lattice']
med_lattice = plot_fills(eps,data,'Lattice',colors[1],light_colors[1])
# sobol plot
eps = np.array(epsilons[2])
data = nt_data['Sobol']
med_sobol = plot_fills(eps,data,'Sobol',colors[2],light_colors[2])
# iid bigO
ax.loglog(eps_iid, (med_iid[0]*eps_iid[0]**2)/(eps_iid**2), '--k', label=r'$\mathcal{O}(1/\epsilon^2)$')
# ld bigO
ax.loglog(eps, ((med_lattice[0]*med_sobol[0])**.5 *eps[0]) / eps , '-.k', label=r'$\mathcal{O}(1/\epsilon)$')
# metas
ax.set_xlabel(r'$\epsilon$')
ax.set_ylabel(label)
ax.spines['right'].set_visible(False)
ax.spines['top'].set_visible(False)
ax.legend(loc='lower left',frameon=False)
ax.set_title(f'Statistics from {trials} runs')
# plot sample data
ax = axs[0]
ax.plot(xplt, yplt, linewidth=lw)
ax.plot(x, y, 'o', markersize=ms, color='orange')
ax.errorbar(x, y, yerr=2 * np.sqrt(v), marker='', linestyle='', color='orange', linewidth=lw)
ax.set_title('Sample data with noise')
ax.set_xlim([0,1])
ax.set_xticks([0,1])
ax.set_ylim([-3, 3])
ax.set_yticks([-3,3]);

