Importance Sampling Examples
from qmcpy import *
from numpy import *
import pandas as pd
pd.options.display.float_format = '{:.2e}'.format
from matplotlib import pyplot as plt
import matplotlib
%matplotlib inline
Game Example
Consider a game where
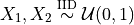 are drawn
with a payoff of
are drawn
with a payoff of
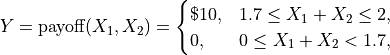
What is the expected payoff of this game?
payoff = lambda x: 10*(x.sum(1)>1.7)
abs_tol = 1e-3
Vanilla Monte Carlo
With ordinary Monte Carlo we do the following:
![\mu = \mathbb{E}(Y) = \int_{[0,1]^2} \text{payoff}(x_1,x_2) \,
\mathrm{d} x_1 \mathrm{d}x_2](../_images/math/adc2df06b15cf8ac84142b29e01db67710c12ff3.png)
d = 2
integral = CustomFun(
true_measure = Uniform(Lattice(d)),
g = payoff)
solution1,data1 = CubQMCLatticeG(integral, abs_tol).integrate()
data1
LDTransformData (AccumulateData Object)
solution 0.450
comb_bound_low 0.449
comb_bound_high 0.450
comb_flags 1
n_total 2^(16)
n 2^(16)
time_integrate 0.043
CubQMCLatticeG (StoppingCriterion Object)
abs_tol 0.001
rel_tol 0
n_init 2^(10)
n_max 2^(35)
CustomFun (Integrand Object)
Uniform (TrueMeasure Object)
lower_bound 0
upper_bound 1
Lattice (DiscreteDistribution Object)
d 2^(1)
dvec [0 1]
randomize 1
order natural
entropy 260768472417330885254602359583380448047
spawn_key ()
Monte Carlo with Importance Sampling
We may add the importance sampling to increase the number of samples with positive payoffs. Let
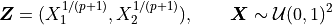
This means that 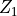 and
and 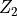 are IID with common CDF
are IID with common CDF
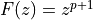 and common PDF
and common PDF 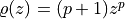 .
Thus,
.
Thus,
![\mu = \mathbb{E}(Y) = \int_{[0,1]^2} \frac{\text{payoff}(z_1,z_2)}{(p+1)^2(z_1z_2)^{p}} \, \varrho(z_1)
\varrho(z_2) \, \mathrm{d} z_1 \mathrm{d}z_2 \\
= \int_{[0,1]^2}
\frac{\text{payoff}(x_1^{1/(p+1)},x_2^{1/(p+1)})}{(p+1)^2(x_1x_2)^{p/(p+1)}}
\, \mathrm{d} x_1 \mathrm{d}x_2](../_images/math/c29707dbb0b59193c3ddb9391cd7f98e0ddb6c91.png)
p = 1
d = 2
integral = CustomFun(
true_measure = Uniform(Lattice(d)),
g = lambda x: payoff(x**(1/(p+1))) / ((p+1)**2 * (x.prod(1))**(p/(p+1))))
solution2,data2 = CubQMCLatticeG(integral, abs_tol).integrate()
data2
LDTransformData (AccumulateData Object)
solution 0.451
comb_bound_low 0.450
comb_bound_high 0.451
comb_flags 1
n_total 2^(14)
n 2^(14)
time_integrate 0.019
CubQMCLatticeG (StoppingCriterion Object)
abs_tol 0.001
rel_tol 0
n_init 2^(10)
n_max 2^(35)
CustomFun (Integrand Object)
Uniform (TrueMeasure Object)
lower_bound 0
upper_bound 1
Lattice (DiscreteDistribution Object)
d 2^(1)
dvec [0 1]
randomize 1
order natural
entropy 101794984569737015308869089738144162915
spawn_key ()
print('Imporance Sampling takes %.3f the time and %.3f the samples'%\
(data2.time_integrate/data1.time_integrate,data2.n_total/data1.n_total))
Imporance Sampling takes 0.451 the time and 0.250 the samples
Asian Call Option Example
The stock price must raise significantly for the payoff to be positive. So we will give an upward drift to the Brownian motion that defines the stock price path. We can think of the option price as the multidimensional integral
![\mu = \mathbb{E}[f(\boldsymbol{X})] = \int_{\mathbb{R}^d}
f(\boldsymbol{x})
\frac{\exp\bigl(-\frac{1}{2} \boldsymbol{x}^T\mathsf{\Sigma}^{-1}
\boldsymbol{x}\bigr)}
{\sqrt{(2 \pi)^{d} \det(\mathsf{\Sigma})}} \, \mathrm{d} \boldsymbol{x}](../_images/math/8c3ec0f4f51d93f6048cefc5928785f5c31703a2.png)
where
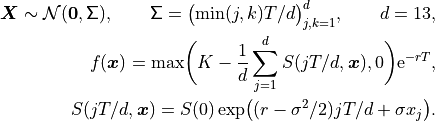
We will replace 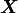 by
by
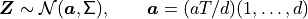
where a positive  will create more positive payoffs. This
corresponds to giving our Brownian motion a drift. To do this we
re-write the integral as
will create more positive payoffs. This
corresponds to giving our Brownian motion a drift. To do this we
re-write the integral as
![\mu = \mathbb{E}[f_{\mathrm{new}}(\boldsymbol{Z})]
= \int_{\mathbb{R}^d}
f_{\mathrm{new}}(\boldsymbol{z})
\frac{\exp\bigl(-\frac{1}{2} (\boldsymbol{z}-\boldsymbol{a})^T
\mathsf{\Sigma}^{-1}
(\boldsymbol{z} - \boldsymbol{a}) \bigr)}
{\sqrt{(2 \pi)^{d} \det(\mathsf{\Sigma})}} \, \mathrm{d} \boldsymbol{z} ,
\\
f_{\mathrm{new}}(\boldsymbol{z}) =
f(\boldsymbol{z})
\frac{\exp\bigl(-\frac{1}{2} \boldsymbol{z}^T
\mathsf{\Sigma}^{-1} \boldsymbol{z} \bigr)}
{\exp\bigl(-\frac{1}{2} (\boldsymbol{z}-\boldsymbol{a})^T
\mathsf{\Sigma}^{-1}
(\boldsymbol{z} - \boldsymbol{a}) \bigr)}
= f(\boldsymbol{z}) \exp\bigl((\boldsymbol{a}/2 - \boldsymbol{z})^T
\mathsf{\Sigma}^{-1}\boldsymbol{a} \bigr)](../_images/math/1d7a80e9501fc67e7507cc1b638e91c9094238e6.png)
Finally note that
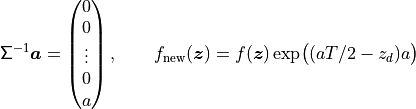
This drift in the Brownian motion may be implemented by changing the
drift input to the BrownianMotion object.
abs_tol = 1e-2
dimension = 32
Vanilla Monte Carlo
integrand = AsianOption(Sobol(dimension))
solution1,data1 = CubQMCSobolG(integrand, abs_tol).integrate()
data1
LDTransformData (AccumulateData Object)
solution 1.788
comb_bound_low 1.782
comb_bound_high 1.794
comb_flags 1
n_total 2^(12)
n 2^(12)
time_integrate 0.014
CubQMCSobolG (StoppingCriterion Object)
abs_tol 0.010
rel_tol 0
n_init 2^(10)
n_max 2^(35)
AsianOption (Integrand Object)
volatility 2^(-1)
call_put call
start_price 30
strike_price 35
interest_rate 0
mean_type arithmetic
dim_frac 0
BrownianMotion (TrueMeasure Object)
time_vec [0.031 0.062 0.094 ... 0.938 0.969 1. ]
drift 0
mean [0. 0. 0. ... 0. 0. 0.]
covariance [[0.031 0.031 0.031 ... 0.031 0.031 0.031]
[0.031 0.062 0.062 ... 0.062 0.062 0.062]
[0.031 0.062 0.094 ... 0.094 0.094 0.094]
...
[0.031 0.062 0.094 ... 0.938 0.938 0.938]
[0.031 0.062 0.094 ... 0.938 0.969 0.969]
[0.031 0.062 0.094 ... 0.938 0.969 1. ]]
decomp_type PCA
Sobol (DiscreteDistribution Object)
d 2^(5)
dvec [ 0 1 2 ... 29 30 31]
randomize LMS_DS
graycode 0
entropy 296048682239793520955192894339316320391
spawn_key ()
Monte Carlo with Importance Sampling
drift = 1
integrand = AsianOption(BrownianMotion(Sobol(dimension),drift=drift))
solution2,data2 = CubQMCSobolG(integrand, abs_tol).integrate()
data2
LDTransformData (AccumulateData Object)
solution 1.783
comb_bound_low 1.776
comb_bound_high 1.790
comb_flags 1
n_total 2^(11)
n 2^(11)
time_integrate 0.014
CubQMCSobolG (StoppingCriterion Object)
abs_tol 0.010
rel_tol 0
n_init 2^(10)
n_max 2^(35)
AsianOption (Integrand Object)
volatility 2^(-1)
call_put call
start_price 30
strike_price 35
interest_rate 0
mean_type arithmetic
dim_frac 0
BrownianMotion (TrueMeasure Object)
time_vec [0.031 0.062 0.094 ... 0.938 0.969 1. ]
drift 0
mean [0. 0. 0. ... 0. 0. 0.]
covariance [[0.031 0.031 0.031 ... 0.031 0.031 0.031]
[0.031 0.062 0.062 ... 0.062 0.062 0.062]
[0.031 0.062 0.094 ... 0.094 0.094 0.094]
...
[0.031 0.062 0.094 ... 0.938 0.938 0.938]
[0.031 0.062 0.094 ... 0.938 0.969 0.969]
[0.031 0.062 0.094 ... 0.938 0.969 1. ]]
decomp_type PCA
transform BrownianMotion (TrueMeasure Object)
time_vec [0.031 0.062 0.094 ... 0.938 0.969 1. ]
drift 1
mean [0.031 0.062 0.094 ... 0.938 0.969 1. ]
covariance [[0.031 0.031 0.031 ... 0.031 0.031 0.031]
[0.031 0.062 0.062 ... 0.062 0.062 0.062]
[0.031 0.062 0.094 ... 0.094 0.094 0.094]
...
[0.031 0.062 0.094 ... 0.938 0.938 0.938]
[0.031 0.062 0.094 ... 0.938 0.969 0.969]
[0.031 0.062 0.094 ... 0.938 0.969 1. ]]
decomp_type PCA
Sobol (DiscreteDistribution Object)
d 2^(5)
dvec [ 0 1 2 ... 29 30 31]
randomize LMS_DS
graycode 0
entropy 144971093458566807495711477416245436939
spawn_key ()
print('Imporance Sampling takes %.3f the time and %.3f the samples'%\
(data2.time_integrate/data1.time_integrate,data2.n_total/data1.n_total))
Imporance Sampling takes 1.009 the time and 0.500 the samples
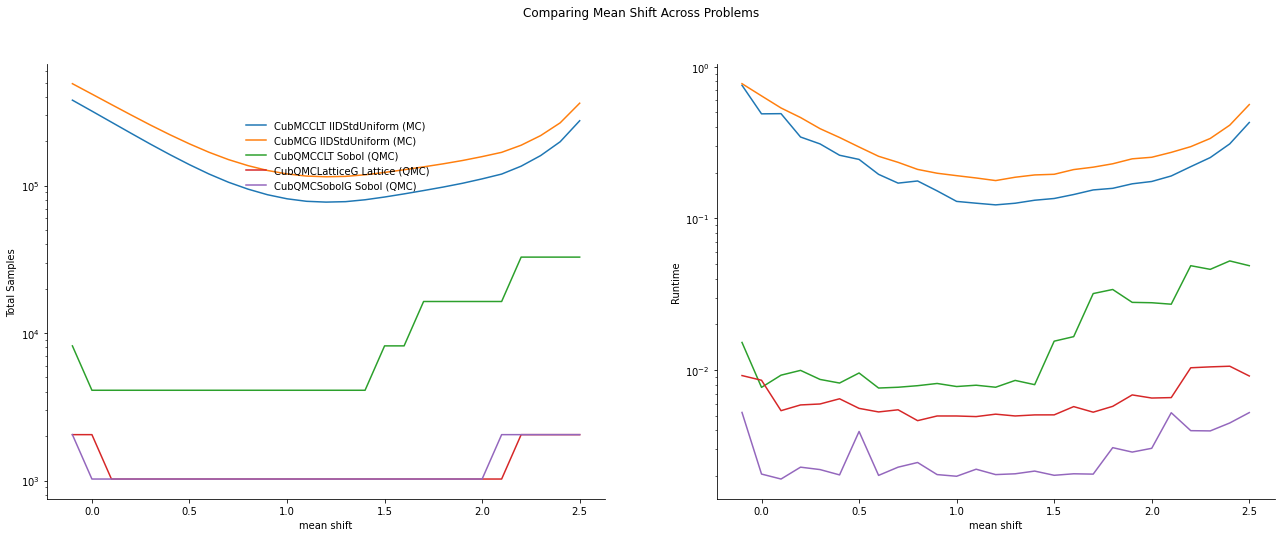
Importance Sampling MC vs QMC
Test Parameters
dimension = 16
abs_tol = .025
trials = 3
df = pd.read_csv('../workouts/mc_vs_qmc/out/importance_sampling.csv')
df['Problem'] = df['Stopping Criterion'] + ' ' + df['Distribution'] + ' (' + df['MC/QMC'] + ')'
df = df.drop(['Stopping Criterion','Distribution','MC/QMC'],axis=1)
problems = ['CubMCCLT IIDStdUniform (MC)',
'CubMCG IIDStdUniform (MC)',
'CubQMCCLT Sobol (QMC)',
'CubQMCLatticeG Lattice (QMC)',
'CubQMCSobolG Sobol (QMC)']
df = df[df['Problem'].isin(problems)]
mean_shifts = df.mean_shift.unique()
df_samples = df.groupby(['Problem'])['n_samples'].apply(list).reset_index(name='n')
df_times = df.groupby(['Problem'])['time'].apply(list).reset_index(name='time')
df.loc[(df.mean_shift==0) | (df.mean_shift==1)].set_index('Problem')
# Note: mean_shift==0 --> NOT using importance sampling
| mean_shift | solution | n_samples | time | |
|---|---|---|---|---|
| Problem | ||||
| CubMCCLT IIDStdUniform (MC) | 0.00e+00 | 1.80e+00 | 3.20e+05 | 4.90e-01 |
| CubMCCLT IIDStdUniform (MC) | 1.00e+00 | 1.77e+00 | 8.15e+04 | 1.30e-01 |
| CubMCG IIDStdUniform (MC) | 0.00e+00 | 1.80e+00 | 4.18e+05 | 6.43e-01 |
| CubMCG IIDStdUniform (MC) | 1.00e+00 | 1.77e+00 | 1.20e+05 | 1.91e-01 |
| CubQMCCLT Sobol (QMC) | 0.00e+00 | 1.78e+00 | 4.10e+03 | 7.69e-03 |
| CubQMCCLT Sobol (QMC) | 1.00e+00 | 1.78e+00 | 4.10e+03 | 7.79e-03 |
| CubQMCLatticeG Lattice (QMC) | 0.00e+00 | 1.78e+00 | 2.05e+03 | 8.56e-03 |
| CubQMCLatticeG Lattice (QMC) | 1.00e+00 | 1.78e+00 | 1.02e+03 | 4.98e-03 |
| CubQMCSobolG Sobol (QMC) | 0.00e+00 | 1.78e+00 | 1.02e+03 | 2.06e-03 |
| CubQMCSobolG Sobol (QMC) | 1.00e+00 | 1.78e+00 | 1.02e+03 | 1.99e-03 |
fig,ax = plt.subplots(nrows=1, ncols=2, figsize=(20, 6))
idx = arange(len(problems))
width = .35
ax[0].barh(idx+width,log(df.loc[df.mean_shift==0]['n_samples'].values),width)
ax[0].barh(idx,log(df.loc[df.mean_shift==1]['n_samples'].values),width)
ax[1].barh(idx+width,df.loc[df.mean_shift==0]['time'].values,width)
ax[1].barh(idx,df.loc[df.mean_shift==1]['time'].values,width)
fig.suptitle('Importance Sampling Comparison by Stopping Criterion on Asian Call Option')
xlabs = ['log(Samples)','Time']
for i in range(len(ax)):
ax[i].set_xlabel(xlabs[i])
ax[i].spines['top'].set_visible(False)
ax[i].spines['bottom'].set_visible(False)
ax[i].spines['right'].set_visible(False)
ax[i].spines['left'].set_visible(False)
ax[1].legend(['Vanilla Monte Carlo','Importance Sampling\nMean Shift=1'],loc='upper right',frameon=False)
ax[1].get_yaxis().set_ticks([])
ax[0].set_yticks(idx)
ax[0].set_yticklabels(problems)
plt.tight_layout();
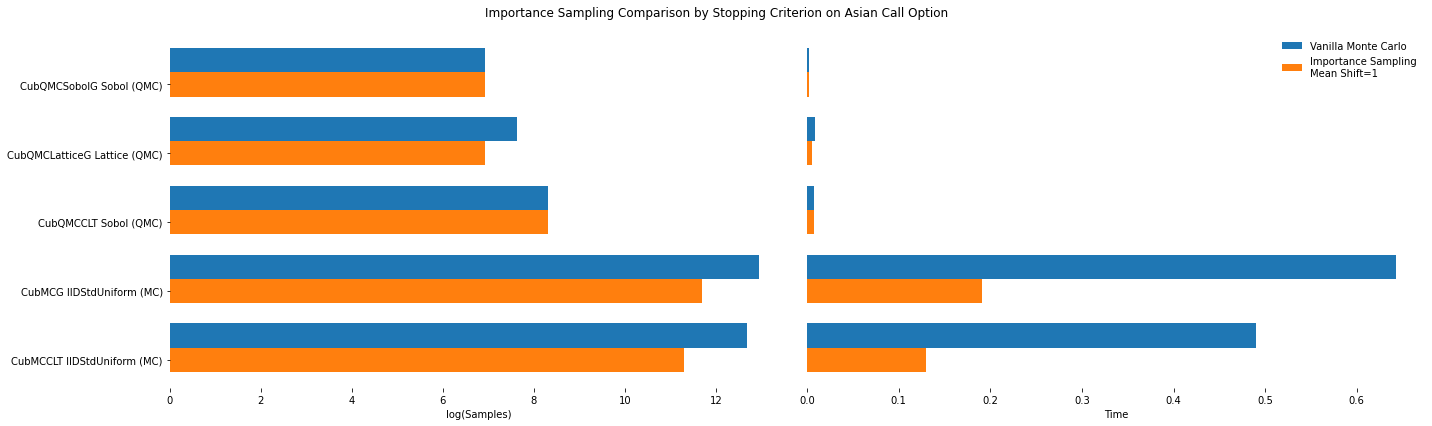
fig,ax = plt.subplots(nrows=1, ncols=2, figsize=(22, 8))
df_samples.apply(lambda row: ax[0].plot(mean_shifts,row.n,label=row['Problem']),axis=1)
df_times.apply(lambda row: ax[1].plot(mean_shifts,row.time,label=row['Problem']),axis=1)
ax[1].legend(frameon=False, loc=(-.85,.7),ncol=1)
ax[0].set_ylabel('Total Samples')
ax[0].set_yscale('log')
ax[1].set_yscale('log')
ax[1].set_ylabel('Runtime')
for i in range(len(ax)):
ax[i].set_xlabel('mean shift')
ax[i].spines['top'].set_visible(False)
ax[i].spines['right'].set_visible(False)
fig.suptitle('Comparing Mean Shift Across Problems');