Gaussian Diagnostics
Experiments to demonstate Guassian assumption used in
cubBayesLattice
import numpy as np
import matplotlib.pyplot as plt
from scipy.optimize import fmin as fminsearch
from numpy import prod, sin, cos, pi
from scipy.stats import norm as gaussnorm
from matplotlib import cm
from qmcpy.integrand import Keister
from qmcpy.discrete_distribution.lattice import Lattice
# print(plt.style.available)
# plt.style.use('./presentation.mplstyle') # use custom settings
plt.style.use('seaborn-poster')
# plt.rcParams.update({'font.size': 12})
# plt.rcParams.update({'lines.linewidth': 2})
# plt.rcParams.update({'lines.markersize': 6})
Let us define the objective function. (cubBayesLattice) finds
optimal parameters by minimizing the objective function
def ObjectiveFunction(theta, order, xun, ftilde):
tol = 100 * np.finfo(float).eps
n = len(ftilde)
arbMean = True
Lambda = kernel2(theta, order, xun)
# compute RKHSnorm
# temp = abs(ftilde(Lambda != 0).^ 2)./ (Lambda(Lambda != 0))
temp = abs(ftilde[Lambda > tol] ** 2) / (Lambda[Lambda > tol])
# compute loss: MLE
if arbMean == True:
RKHSnorm = sum(temp[1:]) / n
temp_1 = sum(temp[1:])
else:
RKHSnorm = sum(temp) / n
temp_1 = sum(temp)
# ignore all zero eigenvalues
loss1 = sum(np.log(Lambda[Lambda > tol])) / n
loss2 = np.log(temp_1)
loss = (loss1 + loss2)
if np.imag(loss) != 0:
# keyboard
raise('error ! : loss value is complex')
# print('L1 %1.3f L2 %1.3f L %1.3f r %1.3e theta %1.3e\n'.format(loss1, loss2, loss, order, theta))
return loss, Lambda, RKHSnorm
Series approximation of the shift invariant kernel
def kernel2(theta, r, xun):
n = xun.shape[0]
m = np.arange(1, (n / 2))
tilde_g_h1 = m ** (-r)
tilde_g = np.hstack([0, tilde_g_h1, 0, tilde_g_h1[::-1]])
g = np.fft.fft(tilde_g)
temp_ = (theta / 2) * g[(xun * n).astype(int)]
C1 = prod(1 + temp_, 1)
# eigenvalues must be real : Symmetric pos definite Kernel
vlambda = np.real(np.fft.fft(C1))
return vlambda
Gaussian random function
def f_rand(xpts, rfun, a, b, c, seed):
dim = xpts.shape[1]
np.random.seed(seed) # initialize random number generator for reproducability
N1 = int(2 ** np.floor(16 / dim))
Nall = N1 ** dim
kvec = np.zeros([dim, Nall]) # initialize kvec
kvec[0, 0:N1] = range(0, N1) # first dimension
Nd = N1
for d in range(1, dim):
Ndm1 = Nd
Nd = Nd * N1
kvec[0:d+1, 0:Nd] = np.vstack([
np.tile(kvec[0:d, 0:Ndm1], (1, N1)),
np.reshape(np.tile(np.arange(0, N1), (Ndm1, 1)), (1, Nd), order="F")
])
kvec = kvec[:, 1: Nall] # remove the zero wavenumber
whZero = np.sum(kvec == 0, axis=0)
abfac = a ** (dim - whZero) * b ** whZero
kbar = np.prod(np.maximum(kvec, 1), axis=0)
totfac = abfac / (kbar ** rfun)
f_c = a * np.random.randn(1, Nall - 1) * totfac
f_s = a * np.random.randn(1, Nall - 1) * totfac
f_0 = c + (b ** dim) * np.random.randn()
argx = np.matmul((2 * np.pi * xpts), kvec)
f_c_ = f_c * np.cos(argx)
f_s_ = f_s * np.sin(argx)
fval = f_0 + np.sum(f_c_ + f_s_, axis=1)
return fval
Periodization transforms
def doPeriodTx(x, integrand, ptransform):
ptransform = ptransform.upper()
if ptransform == 'BAKER': # Baker's transform
xp = 1 - 2 * abs(x - 1 / 2)
w = 1
elif ptransform == 'C0': # C^0 transform
xp = 3 * x ** 2 - 2 * x ** 3
w = prod(6 * x * (1 - x), 1)
elif ptransform == 'C1': # C^1 transform
xp = x ** 3 * (10 - 15 * x + 6 * x ** 2)
w = prod(30 * x ** 2 * (1 - x) ** 2, 1)
elif ptransform == 'C1SIN': # Sidi C^1 transform
xp = x - sin(2 * pi * x) / (2 * pi)
w = prod(2 * sin(pi * x) ** 2, 1)
elif ptransform == 'C2SIN': # Sidi C^2 transform
xp = (8 - 9 * cos(pi * x) + cos(3 * pi * x)) / 16 # psi3
w = prod((9 * sin(pi * x) * pi - sin(3 * pi * x) * 3 * pi) / 16, 1) # psi3_1
elif ptransform == 'C3SIN': # Sidi C^3 transform
xp = (12 * pi * x - 8 * sin(2 * pi * x) + sin(4 * pi * x)) / (12 * pi) # psi4
w = prod((12 * pi - 8 * cos(2 * pi * x) * 2 * pi + sin(4 * pi * x) * 4 * pi) / (12 * pi), 1) # psi4_1
elif ptransform == 'NONE':
xp = x
w = 1
else:
raise (f"The {ptransform} periodization transform is not implemented")
y = integrand(xp) * w
return y
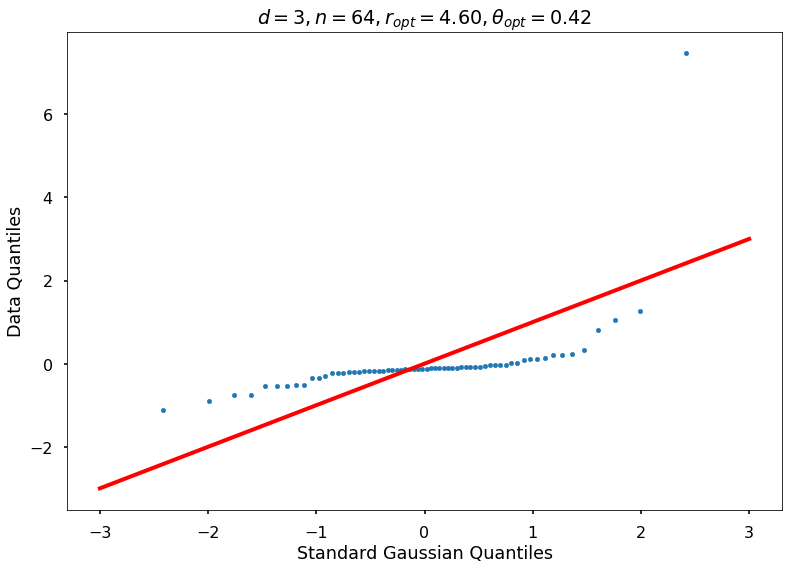
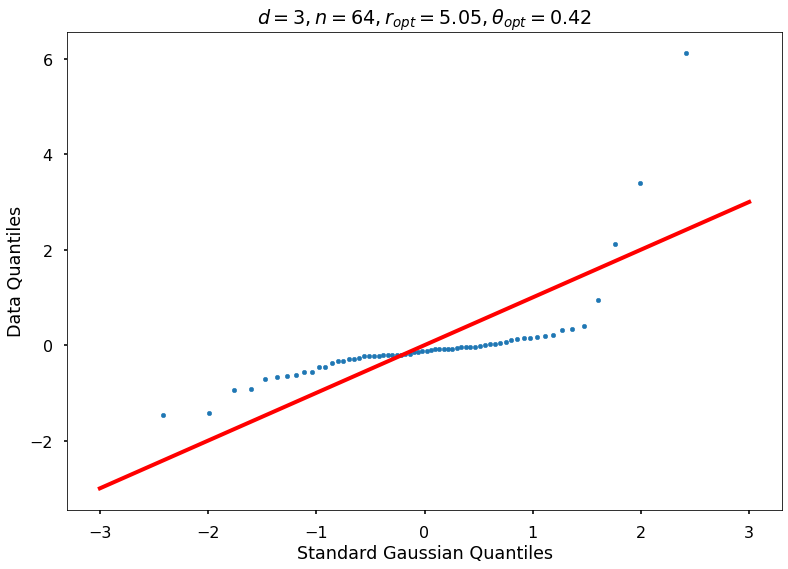
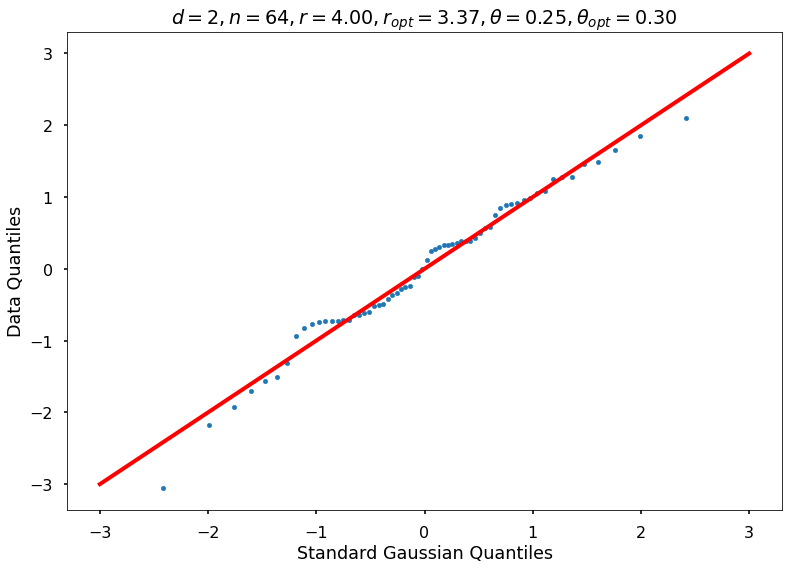
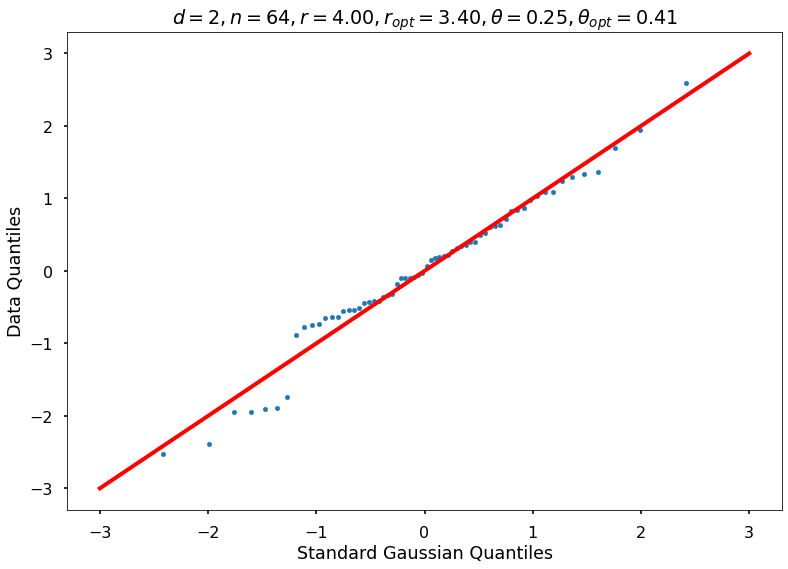
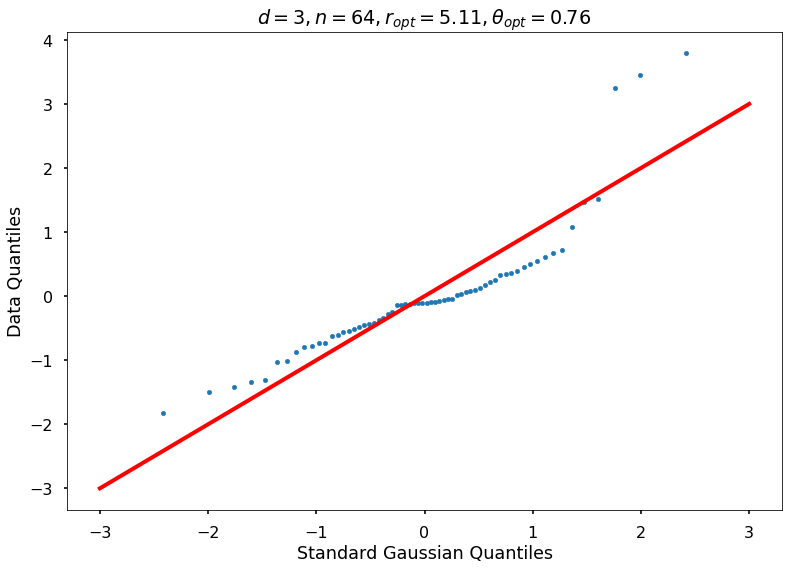
Utility function to draw qqplot or normplot
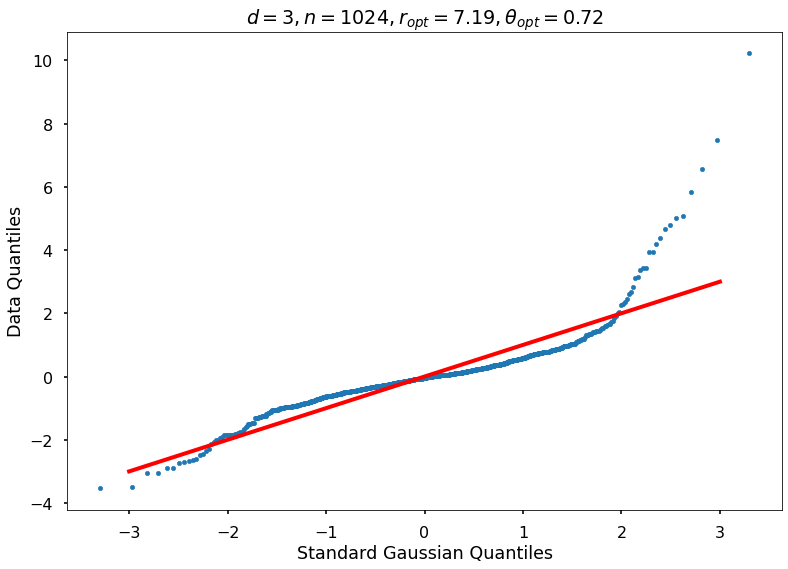
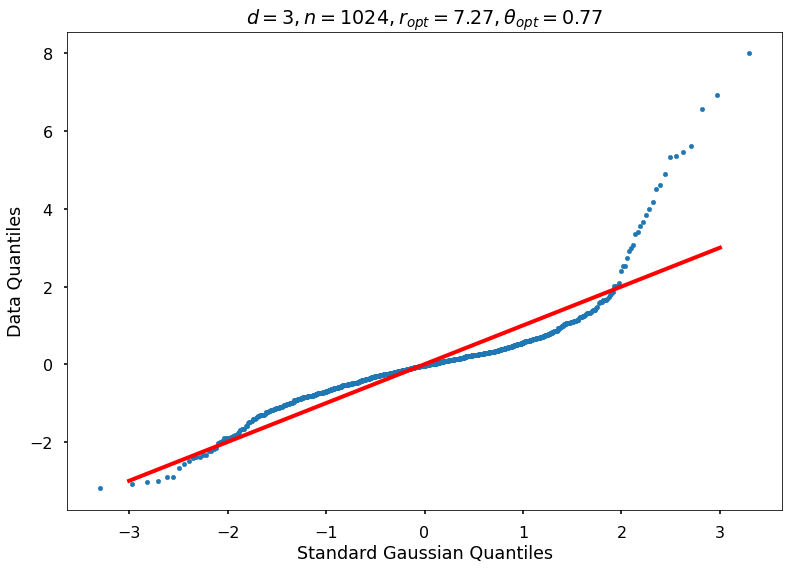
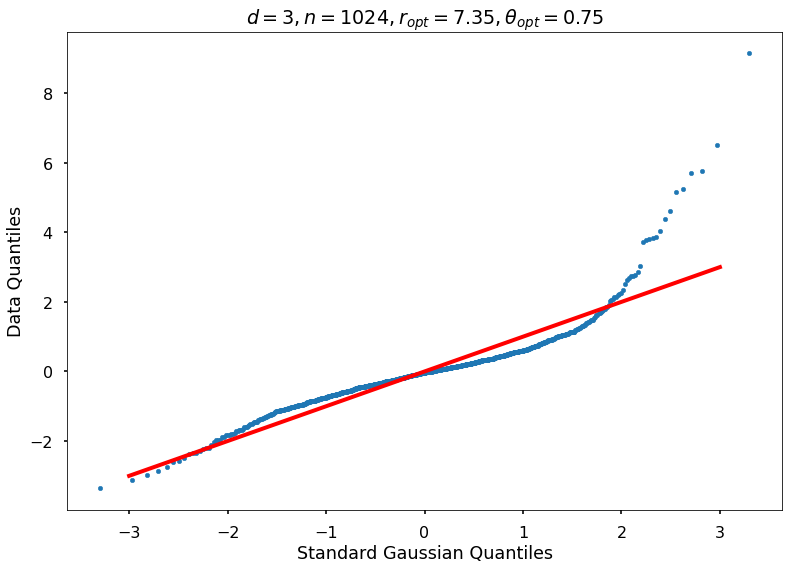
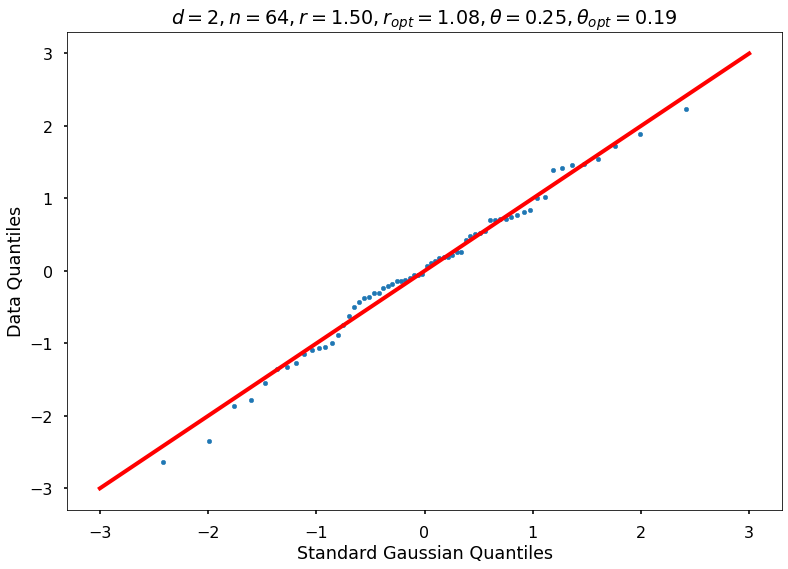
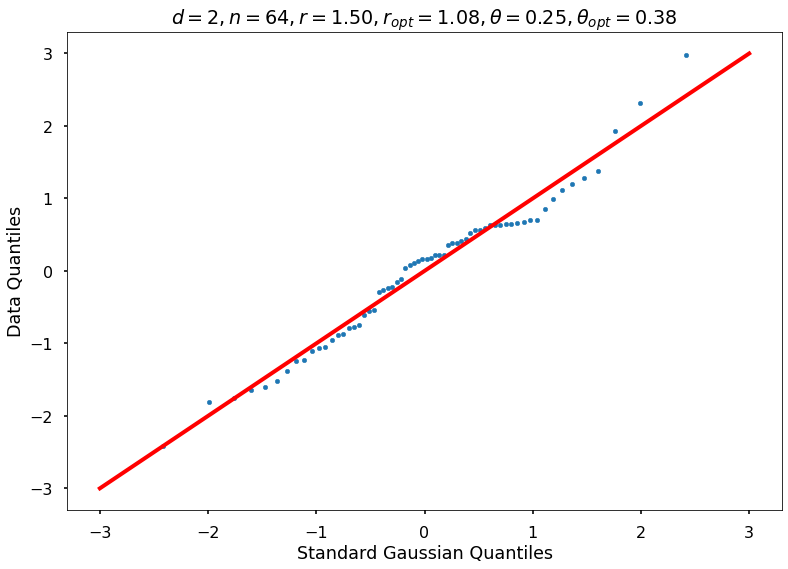
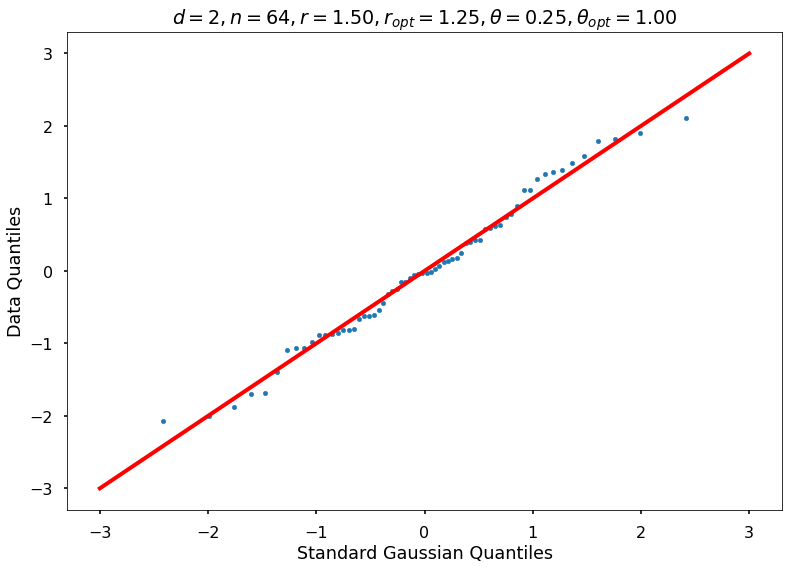
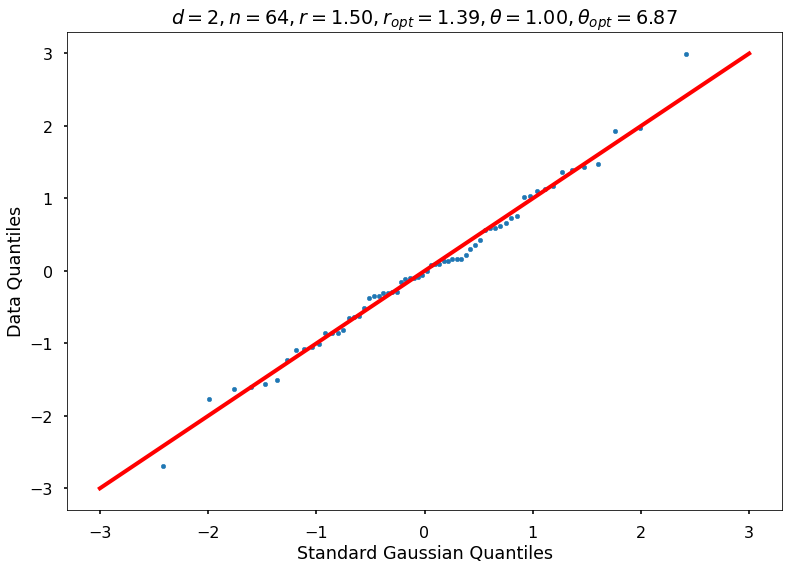
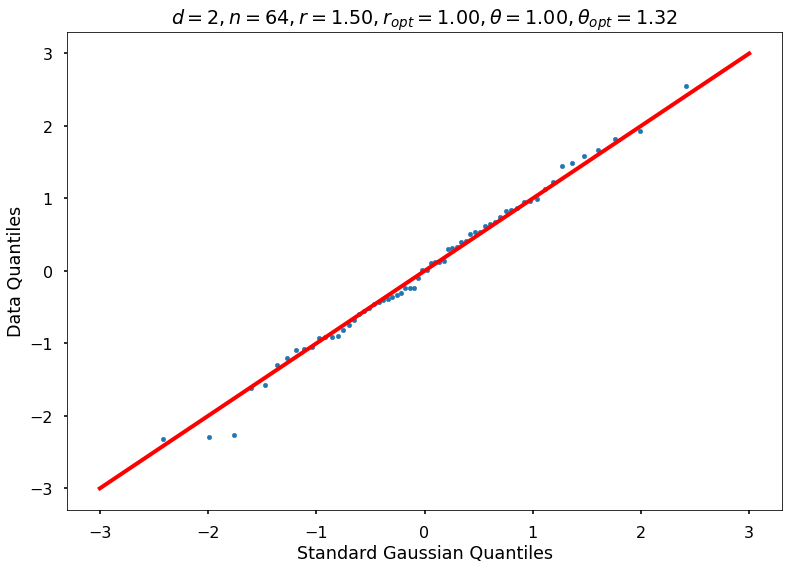
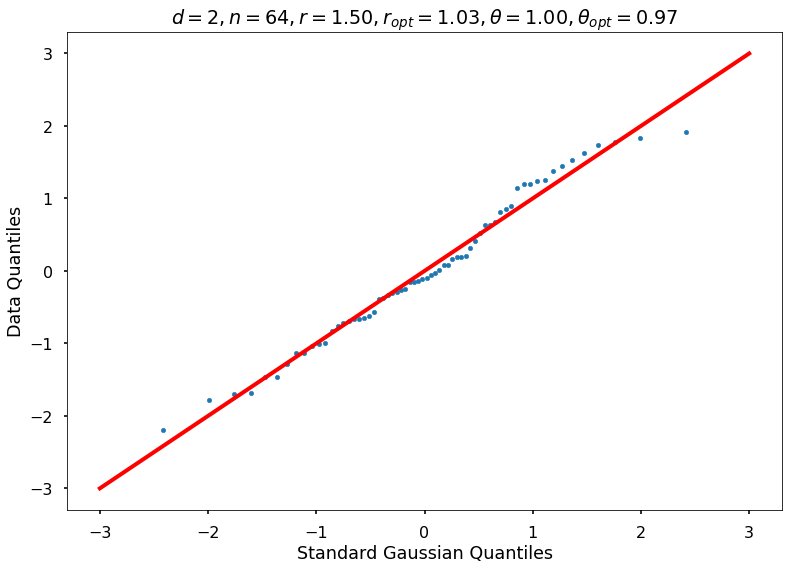
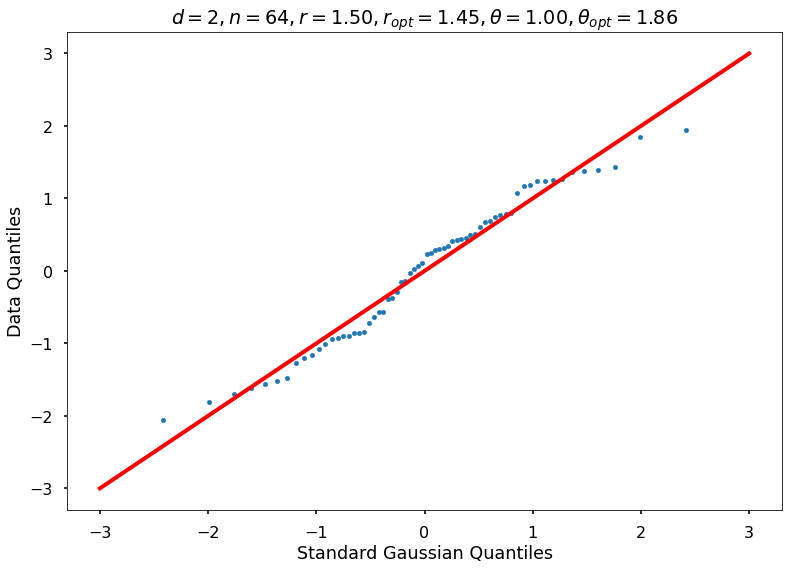
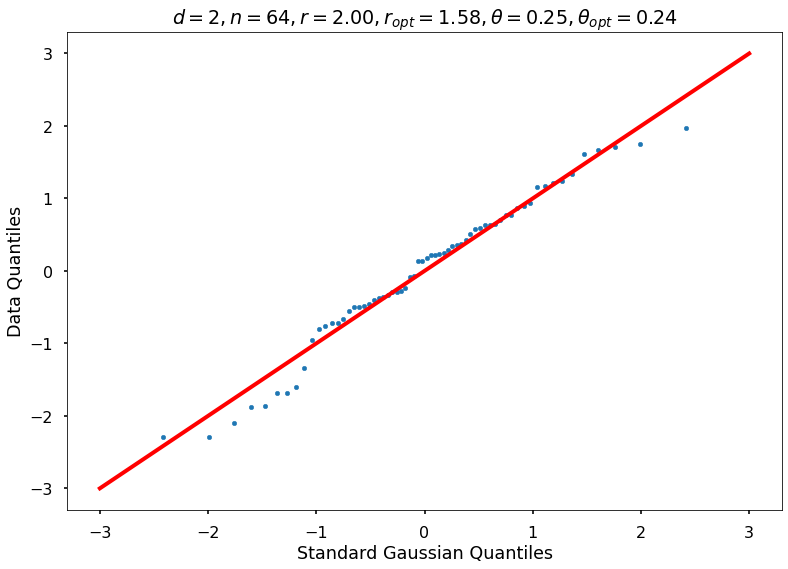
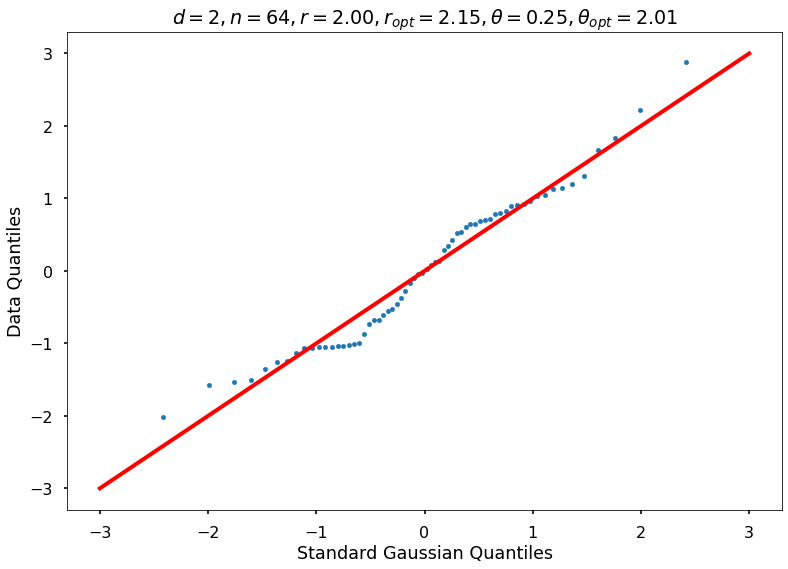
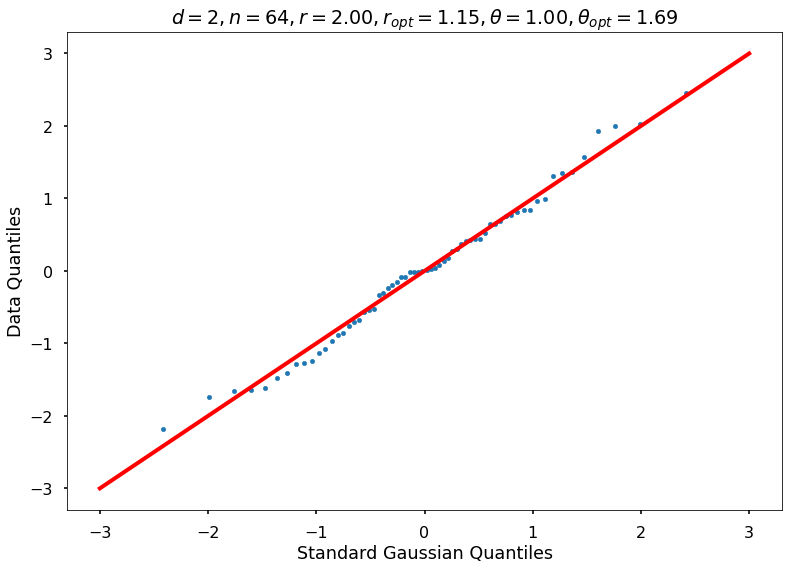
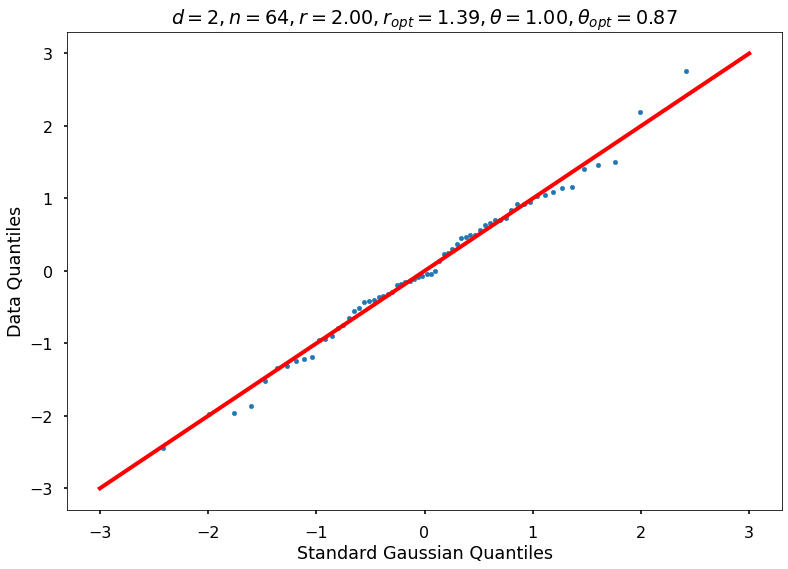
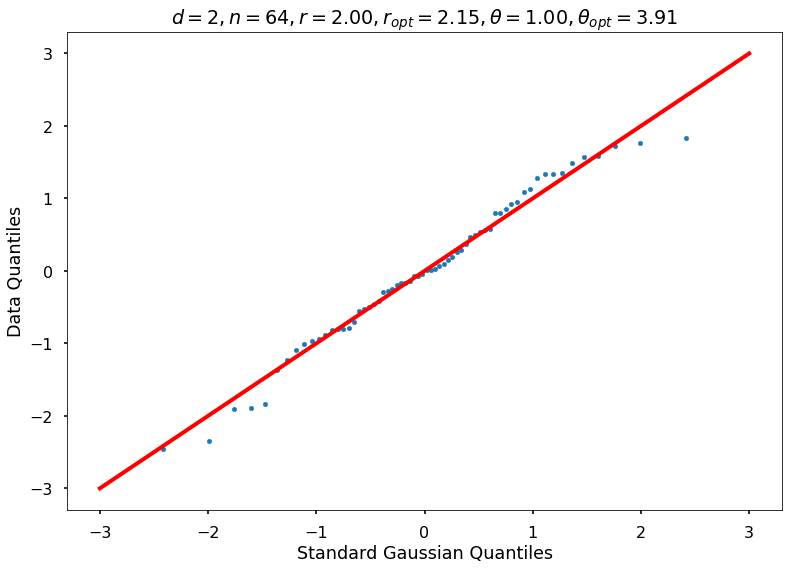
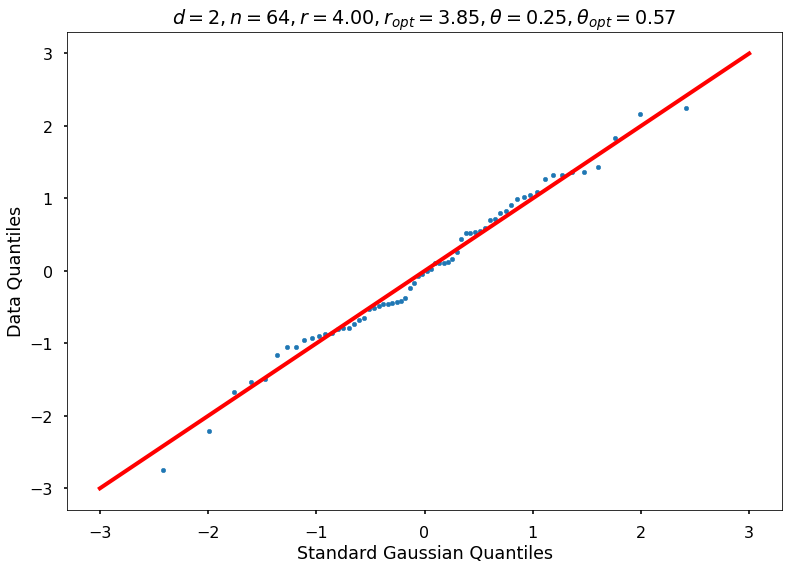
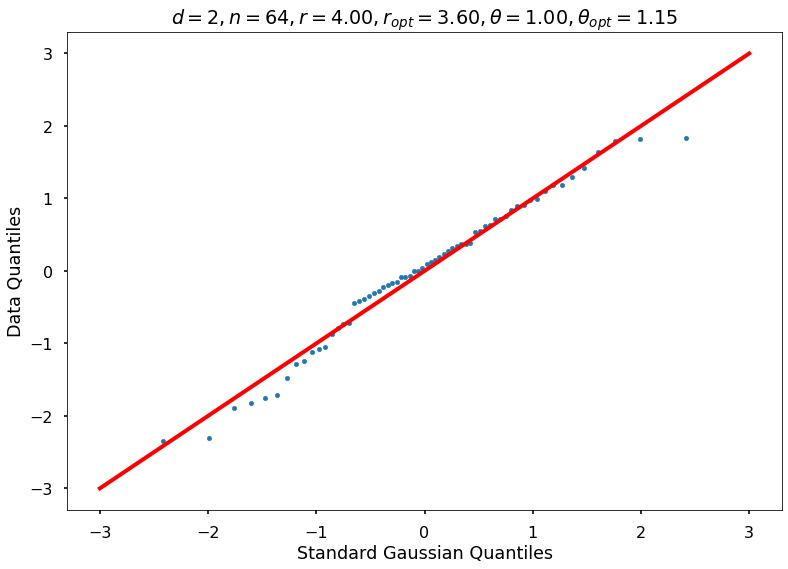
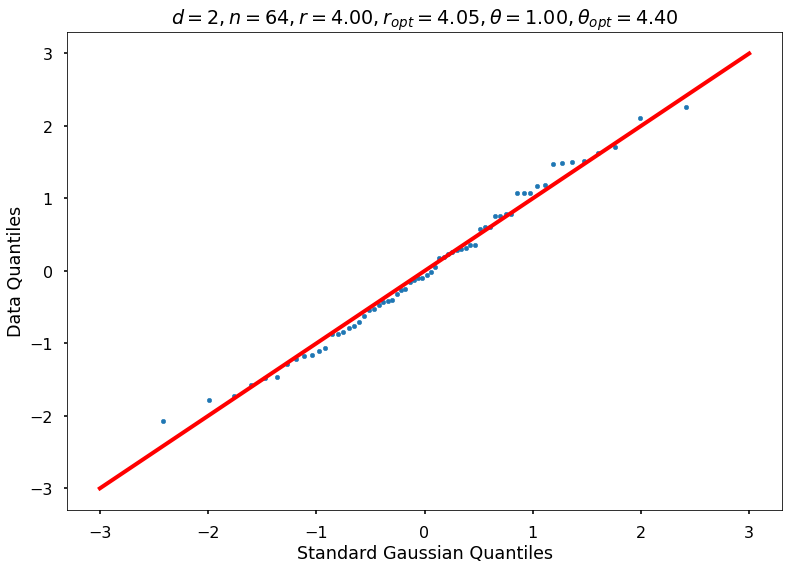
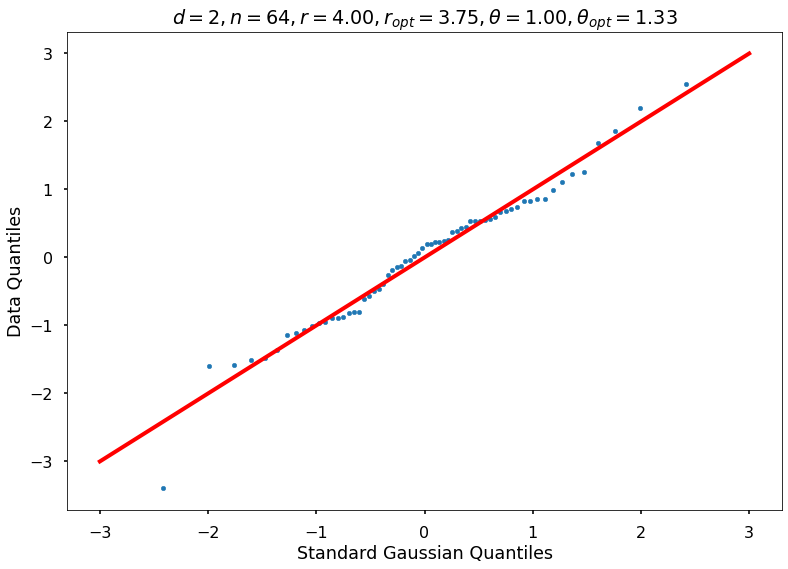
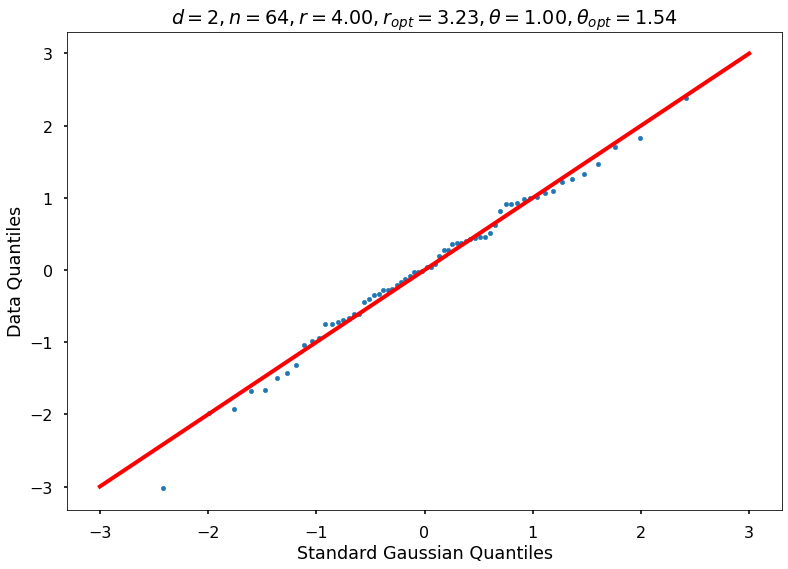
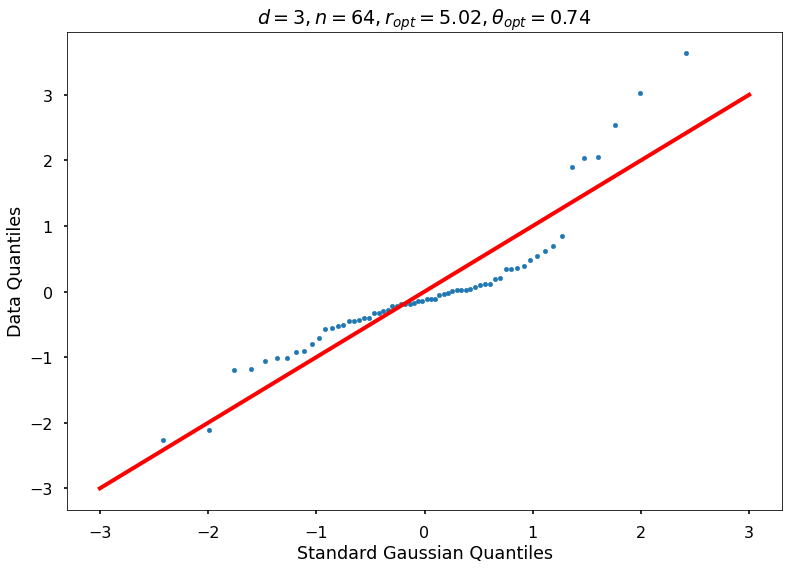
def create_quant_plot(type, vz_real, fName, dim, iii, r, rOpt, theta, thetaOpt):
hFigNormplot, axFigNormplot = plt.subplots()
n = len(vz_real)
if type == 'normplot':
axFigNormplot.normplot(vz_real)
else:
q = (np.arange(1, n + 1) - 1 / 2) / n
stNorm = gaussnorm.ppf(q) # norminv: quantiles of standard normal
axFigNormplot.scatter(stNorm, sorted(vz_real), s=20) # marker='.',
axFigNormplot.plot([-3, 3], [-3, 3], marker='_', linewidth=4, color='red')
axFigNormplot.set_xlabel('Standard Gaussian Quantiles')
axFigNormplot.set_ylabel('Data Quantiles')
if theta:
plt_title = f'$d={dim}, n={n}, r={r:1.2f}, r_{{opt}}={rOpt:1.2f}, \\theta={theta:1.2f}, \\theta_{{opt}}={thetaOpt:1.2f}$'
plt_filename = f'{fName}-QQPlot_n-{n}_d-{dim}_r-{r * 100}_th-{100 * theta}_case-{iii}.jpg'
else:
plt_title = f'$d={dim}, n={n}, r_{{opt}}={rOpt:1.2f}, \\theta_{{opt}}={thetaOpt:1.2f}$'
plt_filename = f'{fName}-QQPlot_n-{n}_d-{dim}_case-{iii}.jpg'
axFigNormplot.set_title(plt_title)
hFigNormplot.savefig(plt_filename)
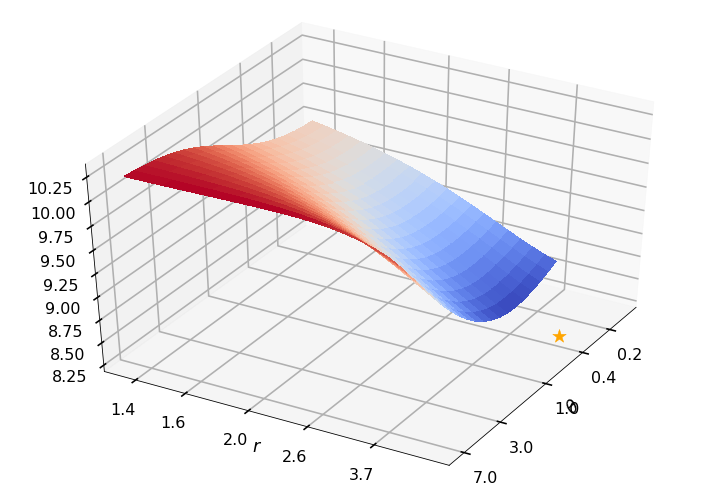
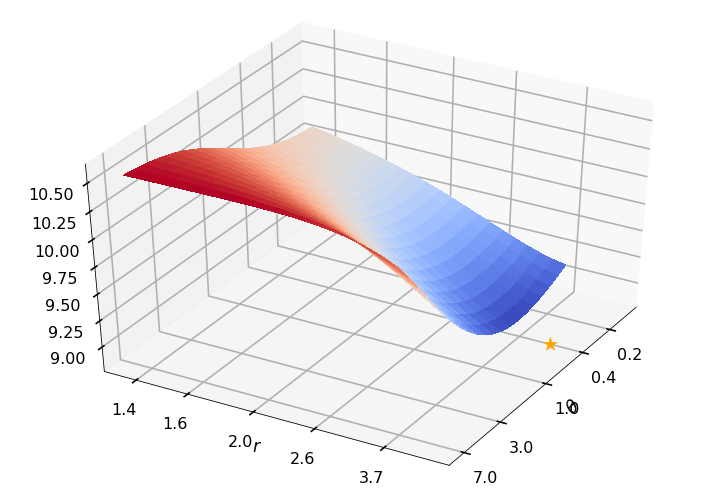
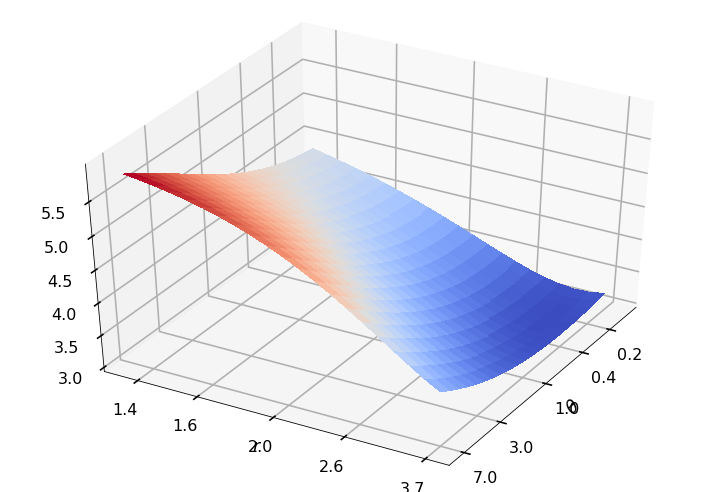
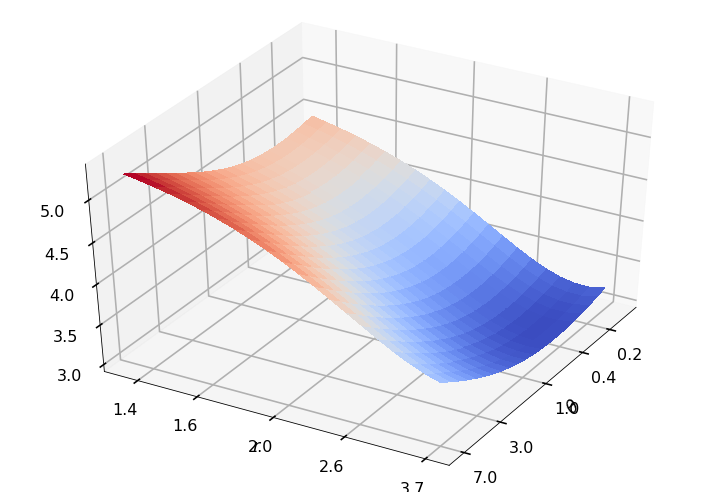
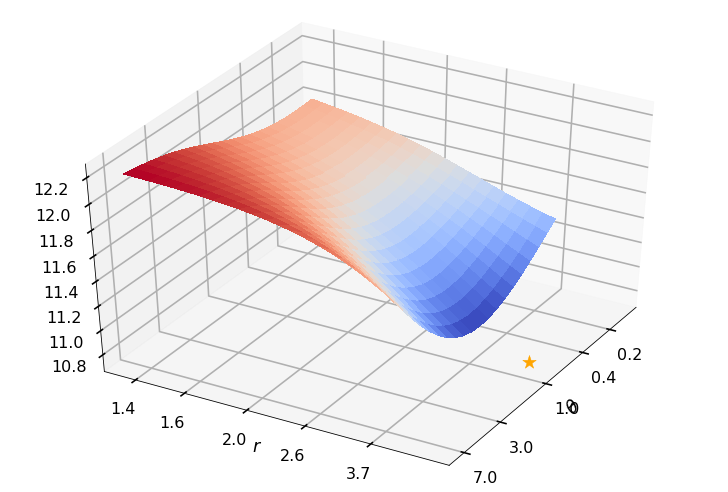
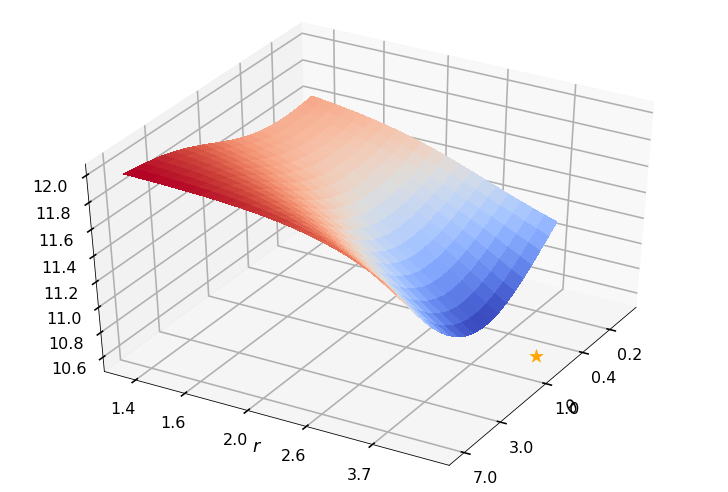
Utility function to plot the objective function and minimum
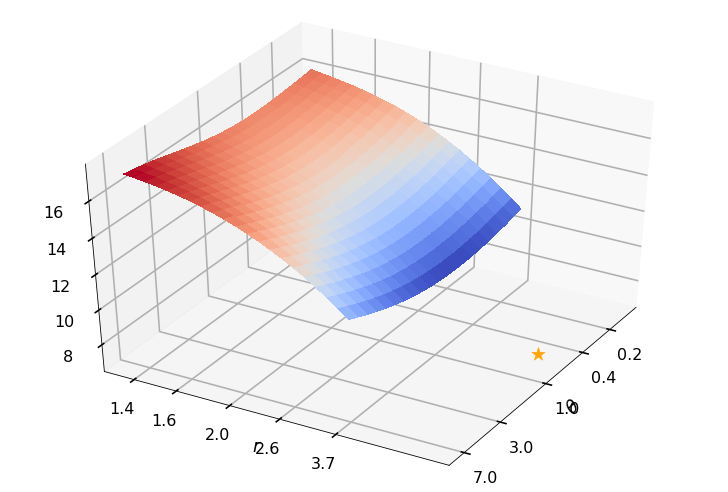
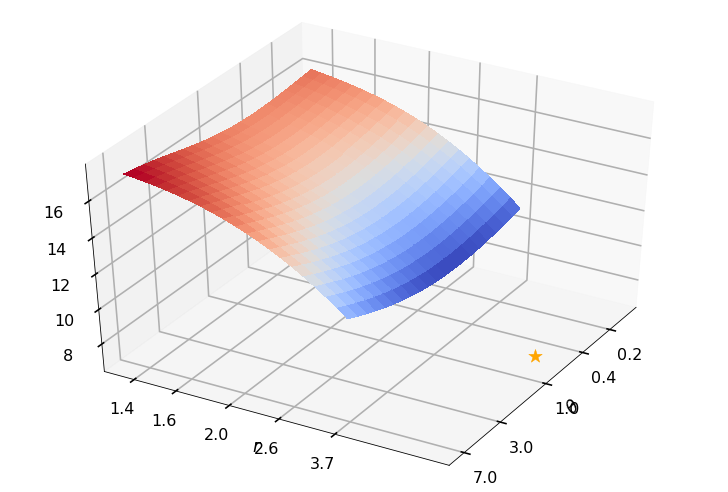
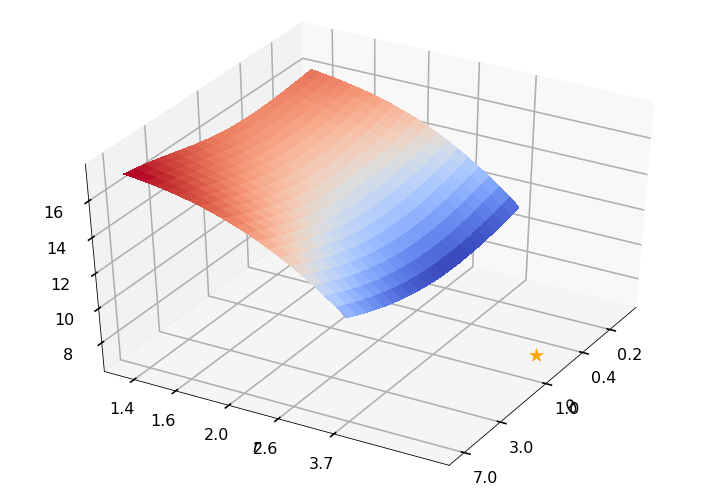
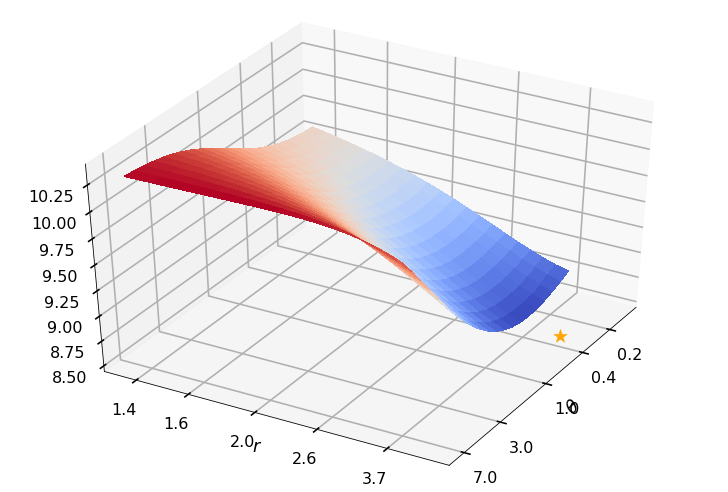
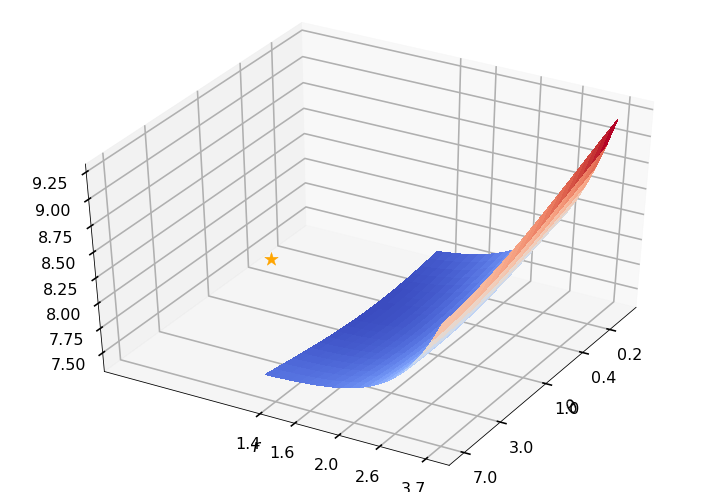
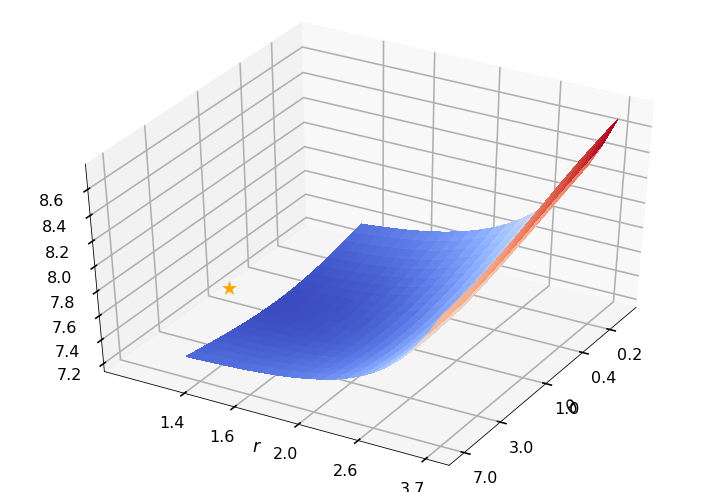
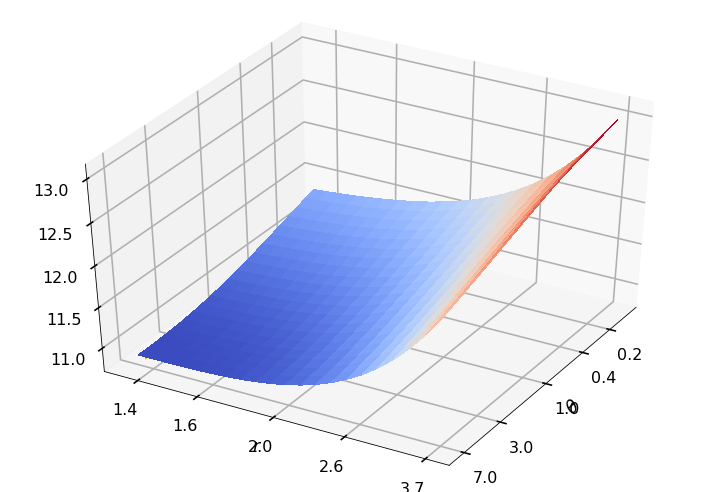
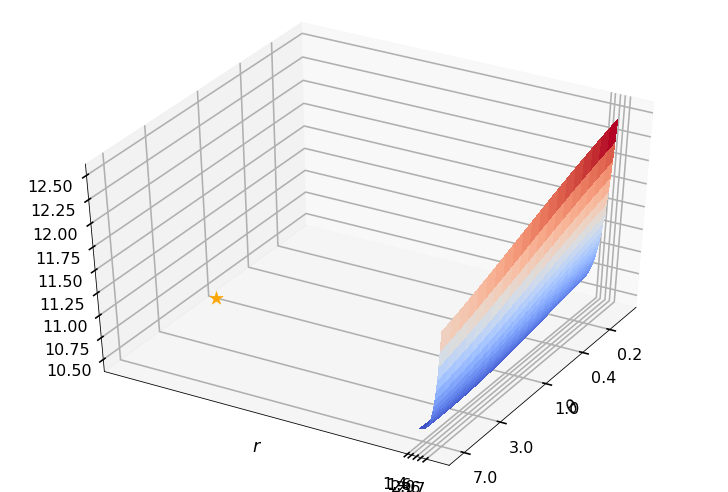
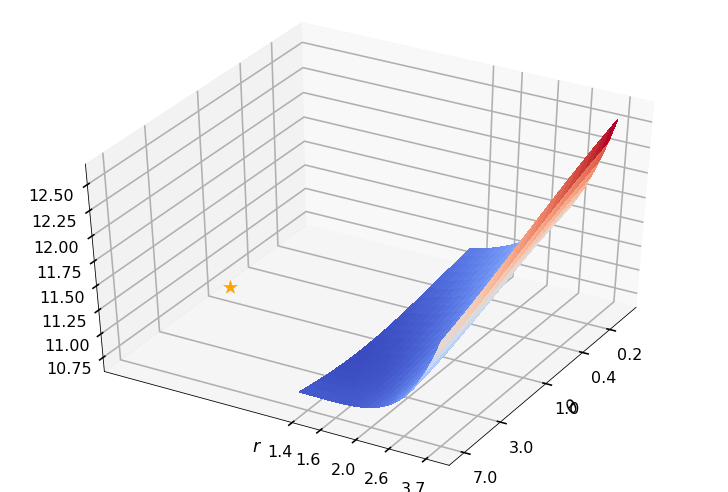
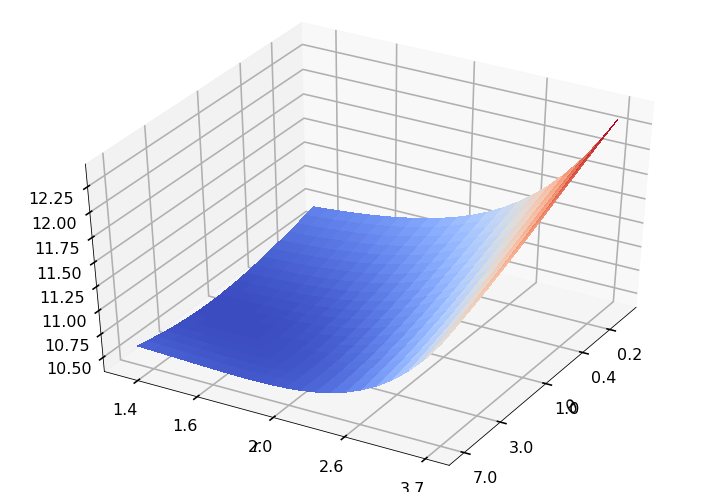
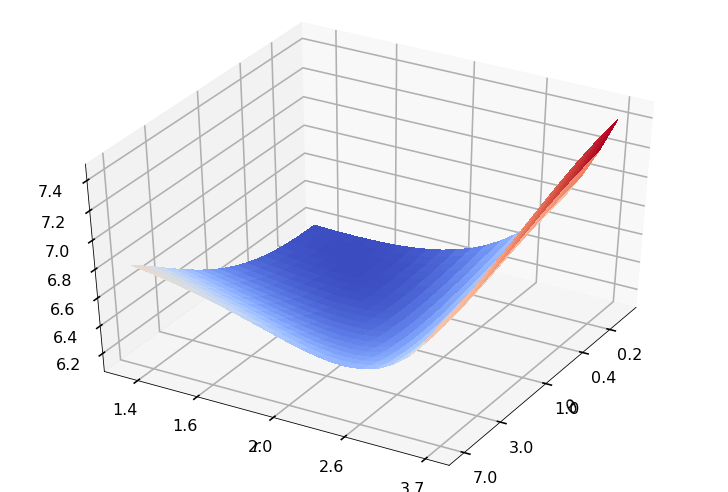
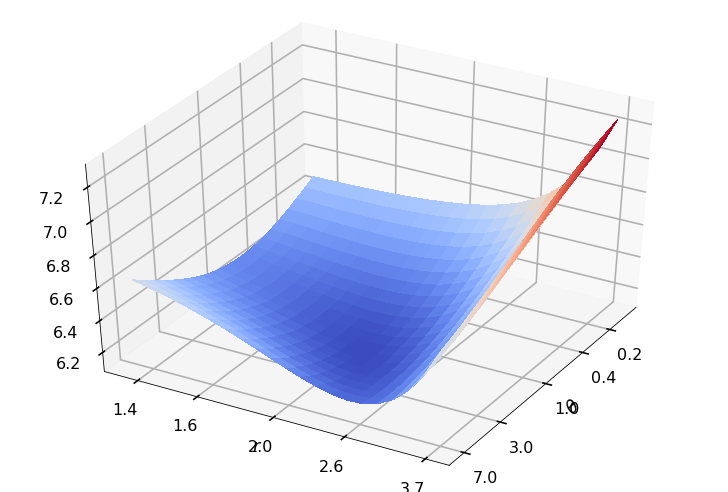
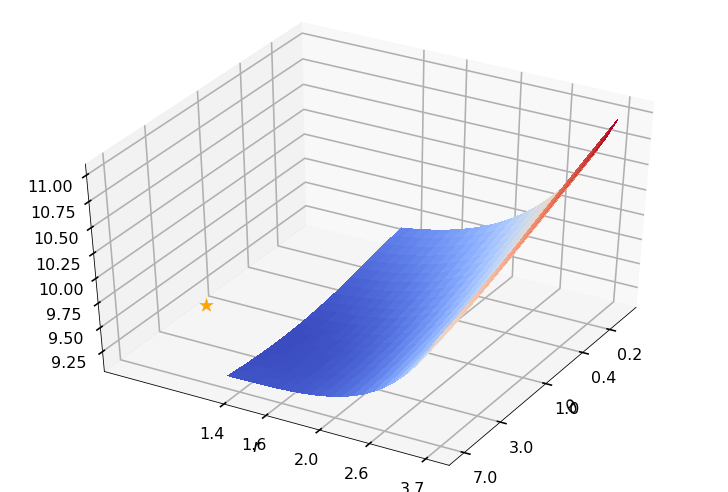
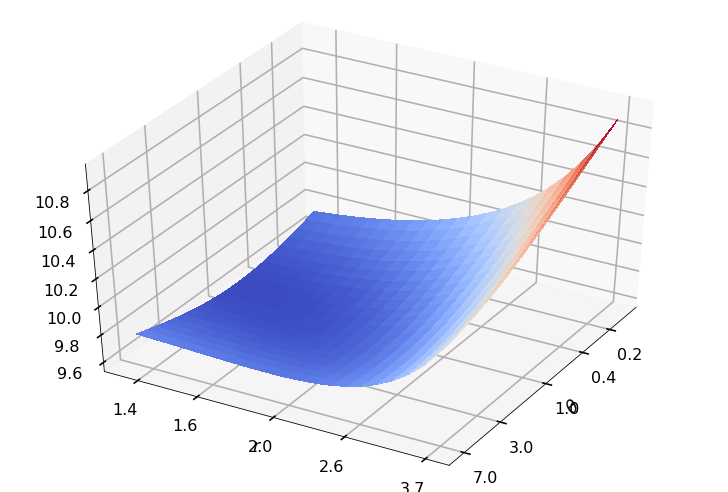
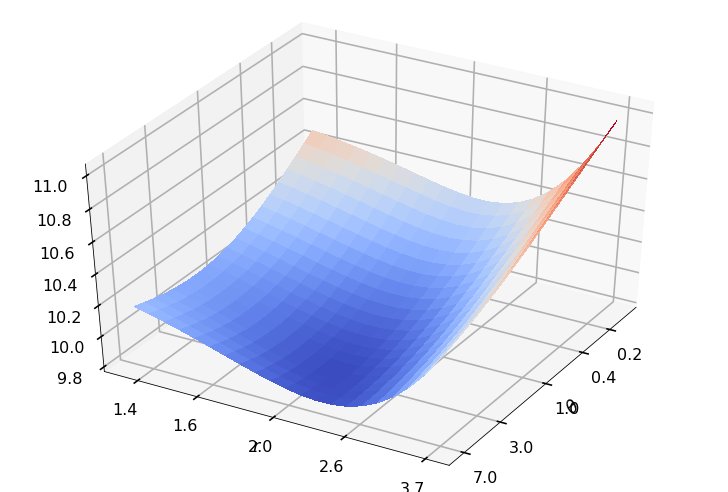
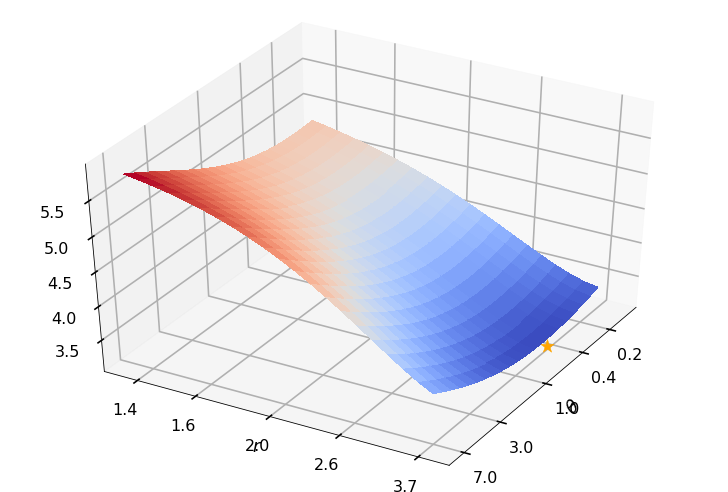
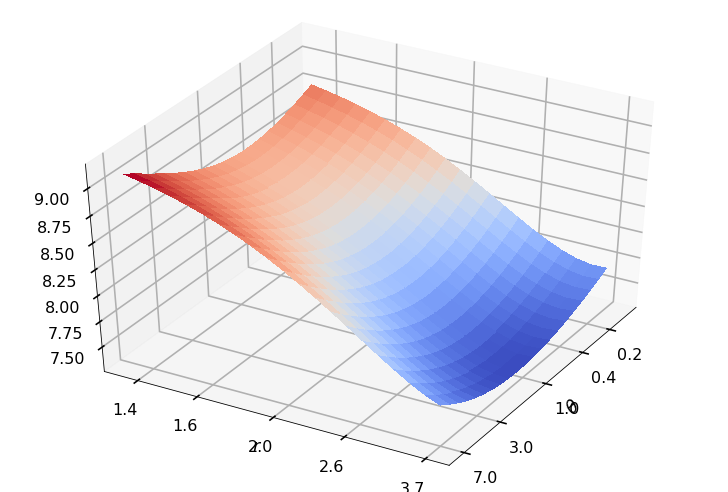
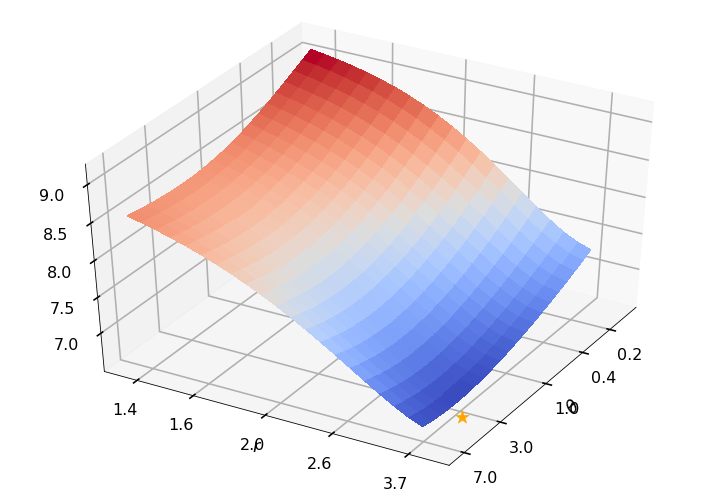
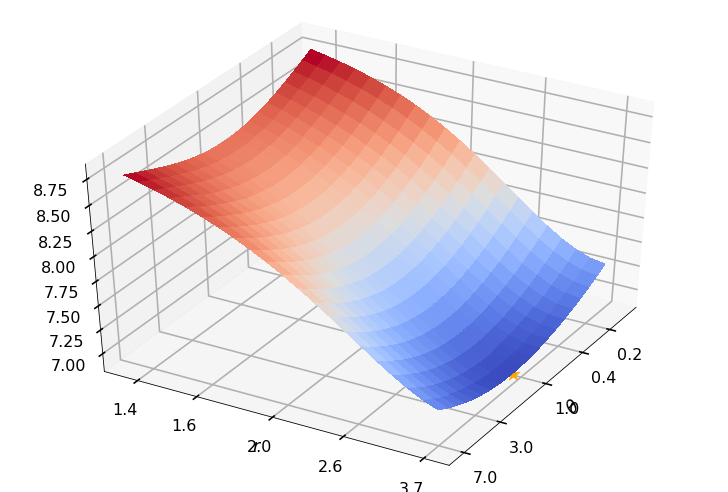
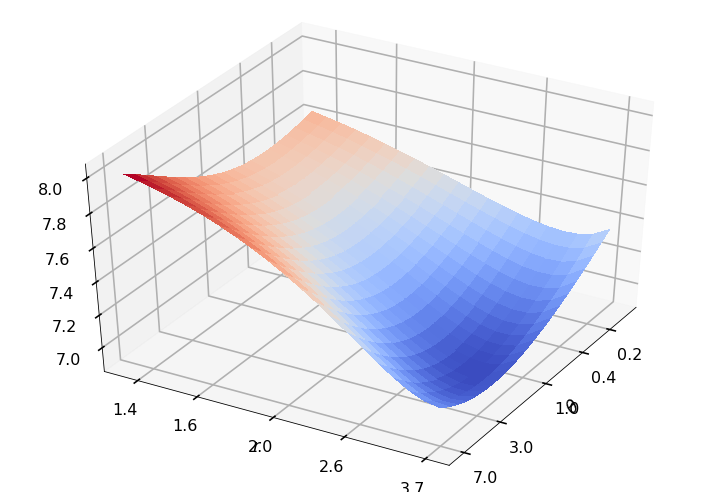
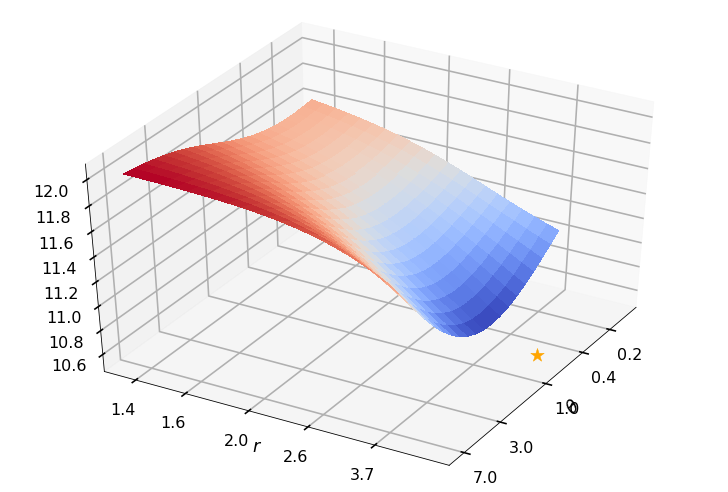
def create_surf_plot(fName, lnthth, lnordord, objfun, objobj, lnParamsOpt, r, theta, iii):
figH, axH = plt.subplots(subplot_kw={"projection": "3d"})
axH.view_init(40, 30)
shandle = axH.plot_surface(lnthth, lnordord, objobj, cmap=cm.coolwarm,
linewidth=0, antialiased=False, alpha=0.8)
xt = np.array([.2, 0.4, 1, 3, 7])
axH.set_xticks(np.log(xt))
axH.set_xticklabels(xt.astype(str))
yt = np.array([1.4, 1.6, 2, 2.6, 3.7])
axH.set_yticks(np.log(yt - 1))
axH.set_yticklabels(yt.astype(str))
axH.set_xlabel('$\\theta$')
axH.set_ylabel('$r$')
axH.scatter(lnParamsOpt[0], lnParamsOpt[1], objfun(lnParamsOpt) * 1.002,
s=200, color='orange', marker='*', alpha=0.8)
if theta:
filename = f'{fName}-ObjFun_n-{npts}_d-{dim}_r-{r * 100}_th-{100 * theta}_case-{iii}.jpg'
else:
filename = f'{fName}-ObjFun_n-{npts}_d-{dim}_case-{iii}.jpg'
figH.savefig(filename)
Minimum working example to demonstrate Gaussian diagnostics concept
def gaussian_diagnostics_engine(whEx, dim, npts, r, fpar, nReps, nPlots):
whEx = whEx - 1
fNames = ['ExpCos', 'Keister', 'rand']
ptransforms = ['none', 'C1sin', 'none']
fName = fNames[whEx]
ptransform = ptransforms[whEx]
rOptAll = [0]*nRep
thOptAll = [0]*nRep
# parameters for random function
# seed = 202326
if whEx == 2:
rfun = r / 2
f_mean = fpar[2]
f_std_a = fpar[0] # this is square root of the a in the talk
f_std_b = fpar[1] # this is square root of the b in the talk
theta = (f_std_a / f_std_b) ** 2
else:
theta = None
for iii in range(nReps):
seed = np.random.randint(low=1, high=1e6) # different each rep
shift = np.random.rand(1, dim)
distribution = Lattice(dimension=dim, order='linear')
xpts, xlat = distribution.gen_samples(n_min=0, n_max=npts, warn=False, return_unrandomized=True)
if fName == 'ExpCos':
integrand = lambda x: np.exp(np.sum(np.cos(2 * np.pi * x), axis=1))
elif fName == 'Keister':
keister = Keister(Lattice(dimension=dim, order='linear'))
integrand = lambda x: keister.f(x)
elif fName == 'rand':
integrand = lambda x: f_rand(x, rfun, f_std_a, f_std_b, f_mean, seed)
else:
print('Invalid function name')
return
y = doPeriodTx(xpts, integrand, ptransform)
ftilde = np.fft.fft(y) # fourier coefficients
ftilde[0] = 0 # ftilde = \mV**H(\vf - m \vone), subtract mean
if dim == 1:
hFigIntegrand = plt.figure()
plt.scatter(xpts, y, 10)
plt.title(f'{fName}_n-{npts}_Tx-{ptransform}')
hFigIntegrand.savefig(f'{fName}_n-{npts}_Tx-{ptransform}_rFun-{rfun:1.2f}.png')
def objfun(lnParams):
loss, Lambda, RKHSnorm = ObjectiveFunction(np.exp(lnParams[0]), 1 + np.exp(lnParams[1]), xlat, ftilde)
return loss
## Plot the objective function
lnthetarange = np.arange(-2, 2.2, 0.2) # range of log(theta) for plotting
lnorderrange = np.arange(-1, 1.1, 0.1) # range of log(r) for plotting
[lnthth, lnordord] = np.meshgrid(lnthetarange, lnorderrange)
objobj = np.zeros(lnthth.shape)
for ii in range(lnthth.shape[0]):
for jj in range(lnthth.shape[1]):
objobj[ii, jj] = objfun([lnthth[ii, jj], lnordord[ii, jj]])
objMinAppx, which = objobj.min(), objobj.argmin()
# [whichrow, whichcol] = ind2sub(lnthth.shape, which)
[whichrow, whichcol] = np.unravel_index(which, lnthth.shape)
lnthOptAppx = lnthth[whichrow, whichcol]
thetaOptAppx = np.exp(lnthOptAppx)
lnordOptAppx = lnordord[whichrow, whichcol]
orderOptAppx = 1 + np.exp(lnordOptAppx)
# print(objMinAppx) # minimum objective function by brute force search
## Optimize the objective function
result = fminsearch(objfun, x0=[lnthOptAppx, lnordOptAppx], xtol=1e-3, full_output=True, disp=False)
lnParamsOpt, objMin = result[0], result[1]
# print(objMin) # minimum objective function by Nelder-Mead
thetaOpt = np.exp(lnParamsOpt[0])
rOpt = 1 + np.exp(lnParamsOpt[1])
rOptAll[iii] = rOpt
thOptAll[iii] = thetaOpt
print(f'{iii}: thetaOptAppx={thetaOptAppx:7.5f}, rOptAppx={orderOptAppx:7.5f}, '
f'objMinAppx={objMinAppx:7.5f}, objMin={objMin:7.5f}')
if iii <= nPlots:
create_surf_plot(fName, lnthth, lnordord, objfun, objobj, lnParamsOpt, r, theta, iii)
vlambda = kernel2(thetaOpt, rOpt, xlat)
s2 = sum(abs(ftilde[2:] ** 2) / vlambda[2:]) / (npts ** 2)
vlambda = s2 * vlambda
# apply transform
# $\vZ = \frac 1n \mV \mLambda**{-\frac 12} \mV**H(\vf - m \vone)$
# np.fft also includes 1/n division
vz = np.fft.ifft(ftilde / np.sqrt(vlambda))
vz_real = np.real(vz) # vz must be real as intended by the transformation
if iii <= nPlots:
create_quant_plot('qqplot', vz_real, fName, dim, iii, r, rOpt, theta, thetaOpt)
r_str = f"{r: 7.5f}" if type(r) == float else str(r)
theta_str = f"{theta: 7.5f}" if type(theta) == float else str(theta)
print(f'\t r = {r_str}, rOpt = {rOpt:7.5f}, theta = {theta_str}, thetaOpt = {thetaOpt:7.5f}\n')
return [theta, rOptAll, thOptAll, fName]
Example 1: Exponential of Cosine
fwh = 1
dim = 3
npts = 2 ** 6
nRep = 20
nPlot = 2
[_, rOptAll, thOptAll, fName] = \
gaussian_diagnostics_engine(fwh, dim, npts, None, None, nRep, nPlot)
## Plot Exponential Cosine example
figH = plt.figure()
plt.scatter(rOptAll, thOptAll, s=20, color='blue')
# axis([4 6 0.1 10])
# set(gca,'yscale','log')
plt.title(f'$d = {dim}, n = {npts}$')
plt.xlabel('Inferred $r$')
plt.ylabel('Inferred $\\theta$')
# print(f'{fName}-rthInfer-n-{npts}-d-{dim}')
figH.savefig(f'{fName}-rthInfer-n-{npts}-d-{dim}.jpg')
0: thetaOptAppx=0.36788, rOptAppx=3.71828, objMinAppx=8.62452, objMin=8.55148
r = None, rOpt = 4.60174, theta = None, thetaOpt = 0.41845
1: thetaOptAppx=0.36788, rOptAppx=3.71828, objMinAppx=8.45167, objMin=8.30125
r = None, rOpt = 5.04805, theta = None, thetaOpt = 0.41943
2: thetaOptAppx=0.44933, rOptAppx=3.71828, objMinAppx=8.94756, objMin=8.86498
r = None, rOpt = 4.70102, theta = None, thetaOpt = 0.53527
3: thetaOptAppx=0.36788, rOptAppx=3.71828, objMinAppx=8.44618, objMin=8.29799
r = None, rOpt = 5.04699, theta = None, thetaOpt = 0.39835
4: thetaOptAppx=0.36788, rOptAppx=3.71828, objMinAppx=8.45494, objMin=8.28315
r = None, rOpt = 5.15814, theta = None, thetaOpt = 0.40447
5: thetaOptAppx=0.36788, rOptAppx=3.71828, objMinAppx=8.57968, objMin=8.47859
r = None, rOpt = 4.75680, theta = None, thetaOpt = 0.38761
6: thetaOptAppx=0.36788, rOptAppx=3.71828, objMinAppx=8.69501, objMin=8.61827
r = None, rOpt = 4.63978, theta = None, thetaOpt = 0.44889
7: thetaOptAppx=0.44933, rOptAppx=3.71828, objMinAppx=9.00638, objMin=8.91494
r = None, rOpt = 4.78933, theta = None, thetaOpt = 0.54575
8: thetaOptAppx=0.44933, rOptAppx=3.71828, objMinAppx=9.01994, objMin=8.90784
r = None, rOpt = 4.97296, theta = None, thetaOpt = 0.55376
9: thetaOptAppx=0.44933, rOptAppx=3.71828, objMinAppx=8.79716, objMin=8.67140
r = None, rOpt = 4.96866, theta = None, thetaOpt = 0.50381
10: thetaOptAppx=0.36788, rOptAppx=3.71828, objMinAppx=8.79790, objMin=8.65545
r = None, rOpt = 5.13328, theta = None, thetaOpt = 0.47860
11: thetaOptAppx=0.36788, rOptAppx=3.71828, objMinAppx=8.47457, objMin=8.34622
r = None, rOpt = 4.90876, theta = None, thetaOpt = 0.42187
12: thetaOptAppx=0.36788, rOptAppx=3.71828, objMinAppx=8.49223, objMin=8.39115
r = None, rOpt = 4.75830, theta = None, thetaOpt = 0.39949
13: thetaOptAppx=0.36788, rOptAppx=3.71828, objMinAppx=8.61363, objMin=8.51504
r = None, rOpt = 4.77039, theta = None, thetaOpt = 0.44782
14: thetaOptAppx=0.30119, rOptAppx=3.71828, objMinAppx=8.48461, objMin=8.34370
r = None, rOpt = 5.03099, theta = None, thetaOpt = 0.35658
15: thetaOptAppx=0.44933, rOptAppx=3.71828, objMinAppx=9.03541, objMin=8.96192
r = None, rOpt = 4.65646, theta = None, thetaOpt = 0.56225
16: thetaOptAppx=0.30119, rOptAppx=3.71828, objMinAppx=8.24322, objMin=8.08881
r = None, rOpt = 4.96519, theta = None, thetaOpt = 0.28016
17: thetaOptAppx=0.36788, rOptAppx=3.71828, objMinAppx=8.53428, objMin=8.37148
r = None, rOpt = 5.12928, theta = None, thetaOpt = 0.43230
18: thetaOptAppx=0.44933, rOptAppx=3.71828, objMinAppx=8.95320, objMin=8.88930
r = None, rOpt = 4.57031, theta = None, thetaOpt = 0.53913
19: thetaOptAppx=0.30119, rOptAppx=3.71828, objMinAppx=8.42682, objMin=8.27037
r = None, rOpt = 5.01433, theta = None, thetaOpt = 0.39608
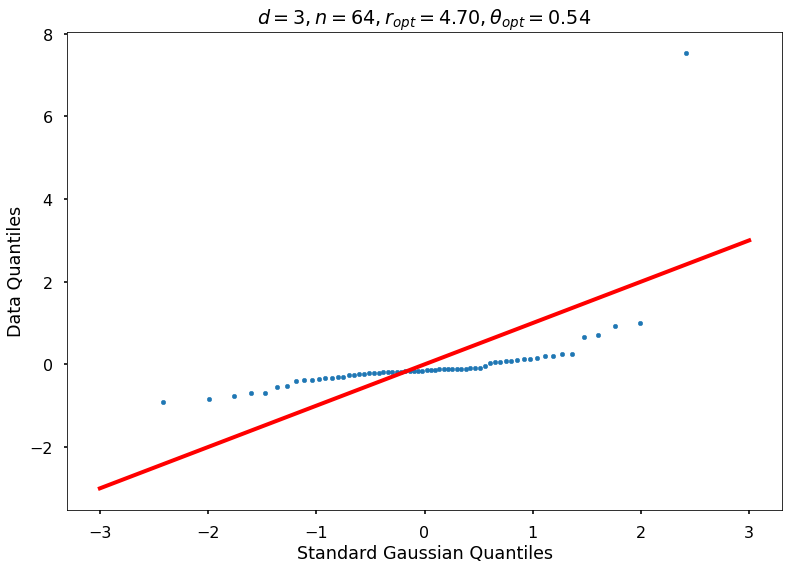
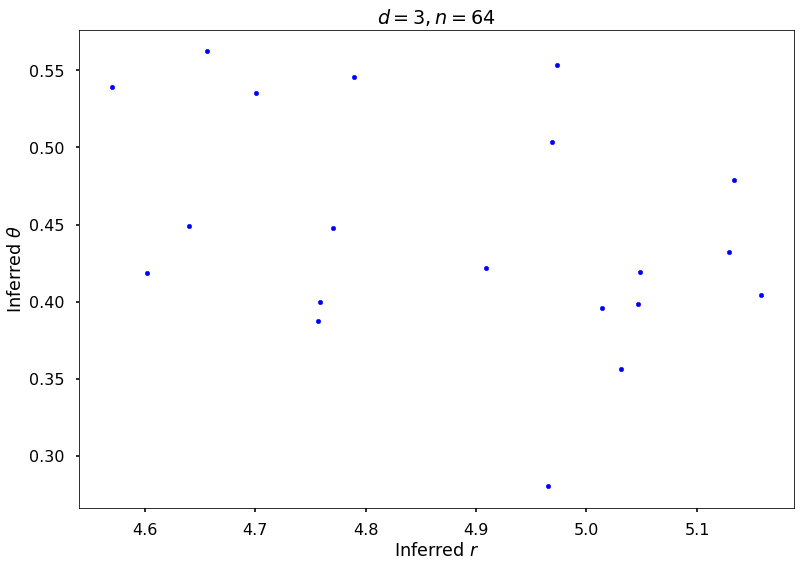
# close all the previous plots to freeup memory
plt.close('all')
Example 2: Random function
## Tests with random function
rArray = [1.5, 2, 4]
nrArr = len(rArray)
fParArray = [[0.5, 1, 2], [1, 1, 1], [1, 1, 1]]
nfPArr = len(fParArray)
fwh = 3
dim = 2
npts = 2 ** 6
nRep = 5 # reduced from 20 to reduce the plots
nPlot = 2
thetaAll = np.zeros((nrArr, nfPArr))
rOptAll = np.zeros((nrArr, nfPArr, nRep))
thOptAll = np.zeros((nrArr, nfPArr, nRep))
for jjj in range(nrArr):
for kkk in range(nfPArr):
thetaAll[jjj, kkk], rOptAll[jjj, kkk, :], thOptAll[jjj, kkk, :], fName = \
gaussian_diagnostics_engine(fwh, dim, npts, rArray[jjj], fParArray[kkk], nRep, nPlot)
7.287181247404372
7.268099510403598
r = 1.50000, rOpt = 1.08321, theta = 0.25000, thetaOpt = 0.18959
7.439206667642468
7.420522178757865
r = 1.50000, rOpt = 1.08048, theta = 0.25000, thetaOpt = 0.37974
7.237565543945999
7.2348776269121124
r = 1.50000, rOpt = 1.24698, theta = 0.25000, thetaOpt = 1.00000
7.144410538581832
7.143842961246502
r = 1.50000, rOpt = 1.75370, theta = 0.25000, thetaOpt = 2.48385
7.317407633559816
7.317303232422838
r = 1.50000, rOpt = 1.55240, theta = 0.25000, thetaOpt = 0.64486
10.864589626523202
10.864398926256648
r = 1.50000, rOpt = 1.38658, theta = 1.00000, thetaOpt = 6.87095
10.506347397179722
10.483539417962092
r = 1.50000, rOpt = 1.00000, theta = 1.00000, thetaOpt = 1.32199
10.732206996328214
10.7195206863197
r = 1.50000, rOpt = 1.03409, theta = 1.00000, thetaOpt = 0.96591
10.713950538487756
10.706754036079342
r = 1.50000, rOpt = 1.70351, theta = 1.00000, thetaOpt = 12.89957
10.3923460325491
10.367674586632736
r = 1.50000, rOpt = 1.00000, theta = 1.00000, thetaOpt = 0.60214
10.481613959193172
10.481573119044445
r = 1.50000, rOpt = 1.44574, theta = 1.00000, thetaOpt = 1.86278
10.624184268139306
10.587513806429946
r = 1.50000, rOpt = 1.00000, theta = 1.00000, thetaOpt = 0.65292
10.336255790040328
10.32105621339534
r = 1.50000, rOpt = 1.11594, theta = 1.00000, thetaOpt = 1.00000
10.699799283646295
10.69967356700936
r = 1.50000, rOpt = 1.44362, theta = 1.00000, thetaOpt = 2.38455
10.51500018761115
10.506939121521446
r = 1.50000, rOpt = 1.17968, theta = 1.00000, thetaOpt = 8.69595
6.158209638152028
6.157963442419079
r = 2, rOpt = 1.57828, theta = 0.25000, thetaOpt = 0.23715
c:toolsminiconda3envsqmcpy1libsite-packagesipykernel_launcher.py:66
RuntimeWarning: More than 20 figures have been opened. Figures created through the pyplot interface (matplotlib.pyplot.figure) are retained until explicitly closed and may consume too much memory. (To control this warning, see the rcParam figure.max_open_warning).
c:toolsminiconda3envsqmcpy1libsite-packagesipykernel_launcher.py:2
RuntimeWarning: More than 20 figures have been opened. Figures created through the pyplot interface (matplotlib.pyplot.figure) are retained until explicitly closed and may consume too much memory. (To control this warning, see the rcParam figure.max_open_warning).
6.157143147450321
6.156543364291681
r = 2, rOpt = 2.15114, theta = 0.25000, thetaOpt = 2.01164
5.75085055457758
5.750809973133201
r = 2, rOpt = 2.00000, theta = 0.25000, thetaOpt = 0.68772
6.069632328951123
6.069055139303397
r = 2, rOpt = 1.84615, theta = 0.25000, thetaOpt = 0.42398
6.034985224985181
6.034874930815441
r = 2, rOpt = 1.67831, theta = 0.25000, thetaOpt = 0.53403
9.551558949940272
9.551384972239866
r = 2, rOpt = 1.92009, theta = 1.00000, thetaOpt = 1.58611
9.633085074190632
9.632776824536194
r = 2, rOpt = 1.76012, theta = 1.00000, thetaOpt = 1.73525
9.568557894047606
9.568348412892643
r = 2, rOpt = 1.58769, theta = 1.00000, thetaOpt = 0.13924
9.541751585509552
9.541667509495124
r = 2, rOpt = 1.48197, theta = 1.00000, thetaOpt = 1.41333
9.5396175595642
9.539398408753463
r = 2, rOpt = 1.55887, theta = 1.00000, thetaOpt = 0.12623
9.170505000843896
9.162768712949173
r = 2, rOpt = 1.15246, theta = 1.00000, thetaOpt = 1.68521
9.6238915085669
9.62377543126308
r = 2, rOpt = 1.39439, theta = 1.00000, thetaOpt = 0.86724
9.851657112032425
9.850539894156423
r = 2, rOpt = 2.15206, theta = 1.00000, thetaOpt = 3.90645
9.806190014351044
9.805541561888338
r = 2, rOpt = 2.44945, theta = 1.00000, thetaOpt = 2.23506
9.79209159625104
9.791515852224858
r = 2, rOpt = 1.85087, theta = 1.00000, thetaOpt = 1.61353
3.2161538975858517
3.211437887705981
r = 4, rOpt = 3.84952, theta = 0.25000, thetaOpt = 0.56819
3.1312343157485762
3.1292745389997414
r = 4, rOpt = 3.37450, theta = 0.25000, thetaOpt = 0.30053
3.0695614957742343
3.0682627340993567
r = 4, rOpt = 3.39840, theta = 0.25000, thetaOpt = 0.41193
3.027380943889609
3.0266075283943112
r = 4, rOpt = 3.76590, theta = 0.25000, thetaOpt = 0.36430
3.5228968345657194
3.5143715614313487
r = 4, rOpt = 3.89398, theta = 0.25000, thetaOpt = 0.48767
7.379762232781738
7.376813143468294
r = 4, rOpt = 3.60031, theta = 1.00000, thetaOpt = 1.14533
6.641928348506947
6.617895389661783
r = 4, rOpt = 4.05057, theta = 1.00000, thetaOpt = 4.39747
7.476855960512737
7.476496711605328
r = 4, rOpt = 3.18967, theta = 1.00000, thetaOpt = 1.00000
6.905066745166625
6.731884736747105
r = 4, rOpt = 4.55762, theta = 1.00000, thetaOpt = 2.69082
6.998288530030136
6.956313755828155
r = 4, rOpt = 4.25120, theta = 1.00000, thetaOpt = 1.18999
6.863019673493637
6.8628173704594655
r = 4, rOpt = 3.43551, theta = 1.00000, thetaOpt = 1.00000
6.942054984148132
6.9414476082169205
r = 4, rOpt = 3.74774, theta = 1.00000, thetaOpt = 1.32587
6.928869540278924
6.928780381246793
r = 4, rOpt = 3.23388, theta = 1.00000, thetaOpt = 1.54465
6.864061134245936
6.84621776419379
r = 4, rOpt = 3.98079, theta = 1.00000, thetaOpt = 1.00000
7.027153534225495
6.917713557698754
r = 4, rOpt = 4.51165, theta = 1.00000, thetaOpt = 2.35318
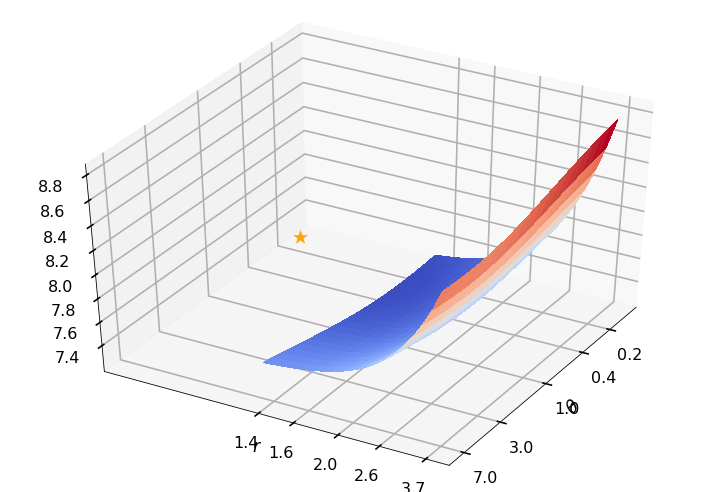
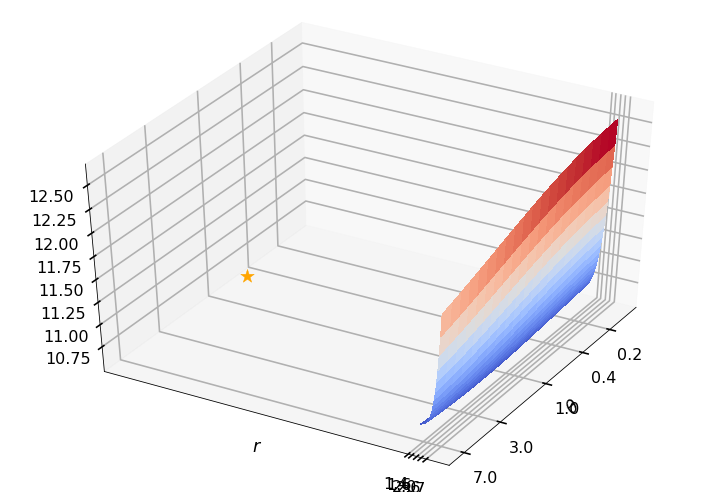
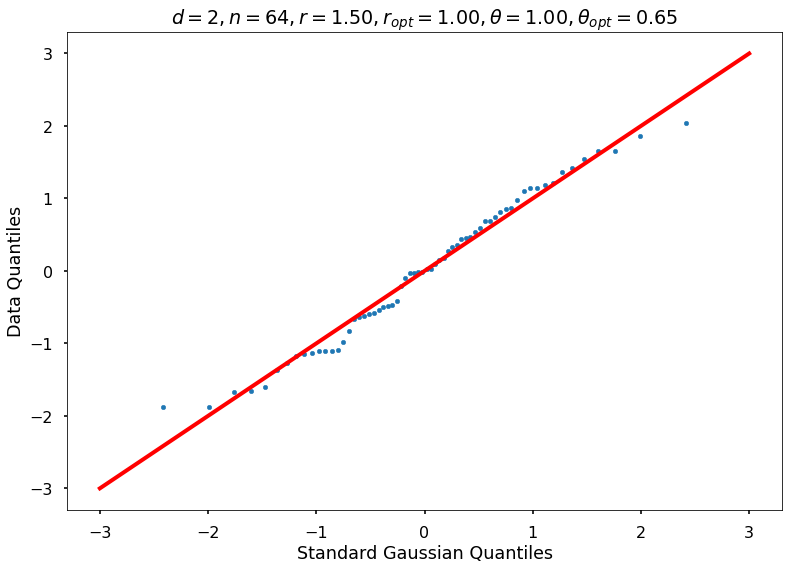
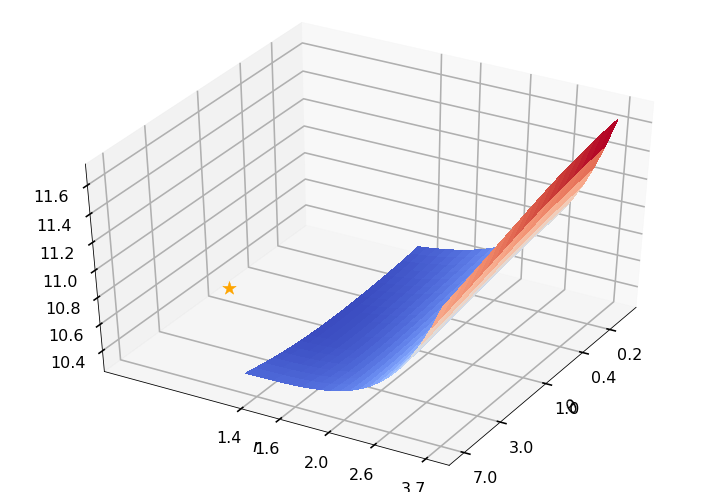
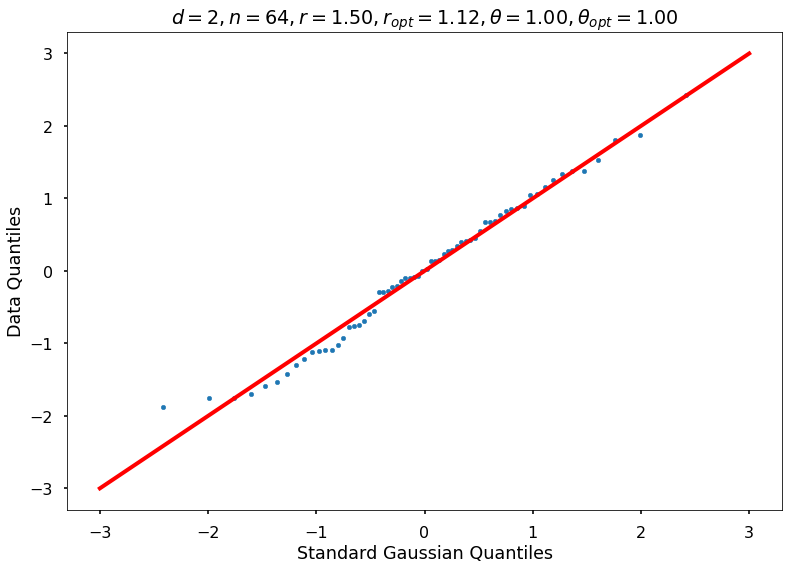
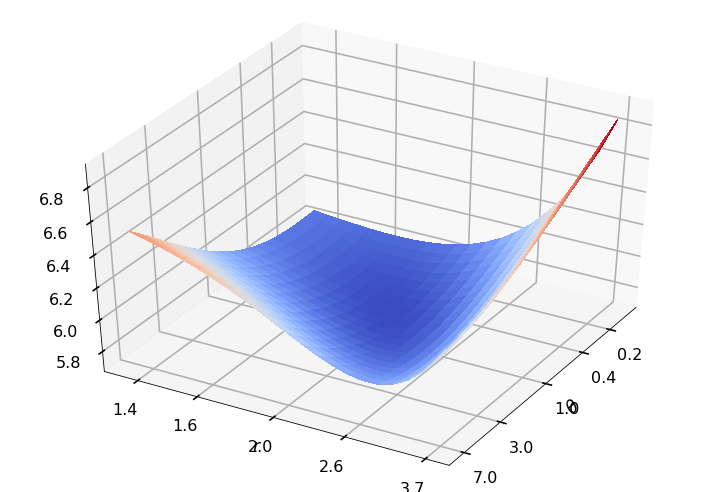
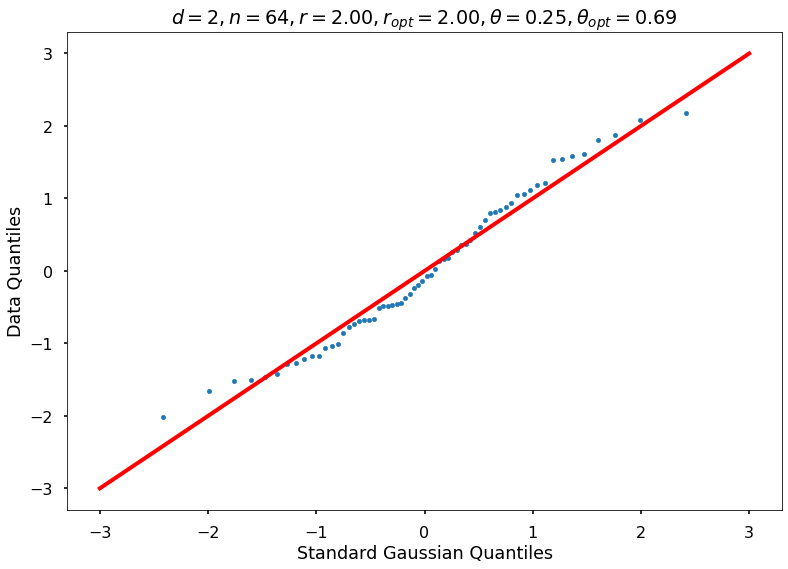
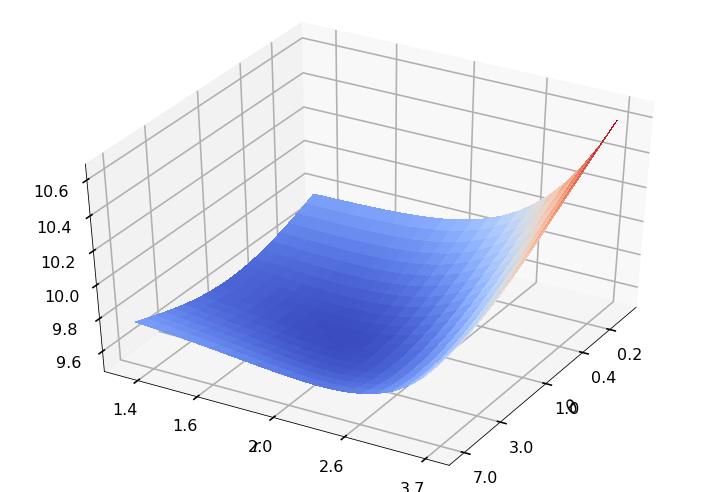
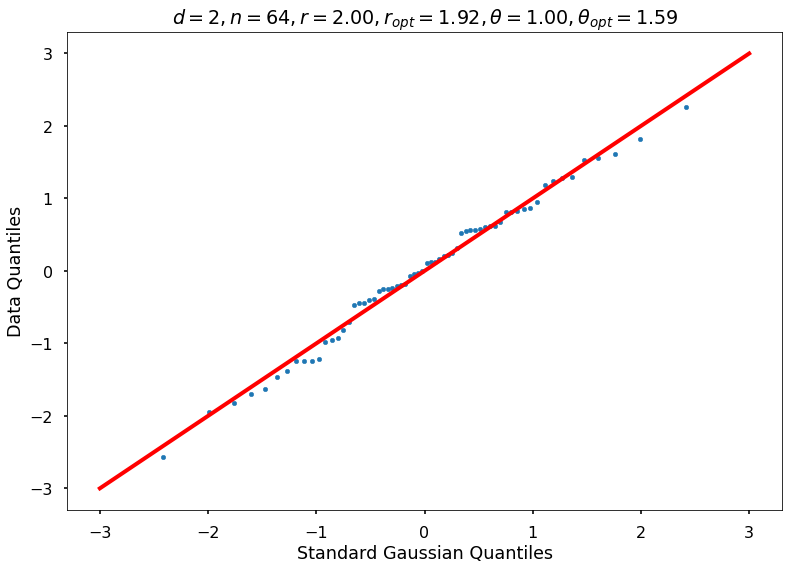
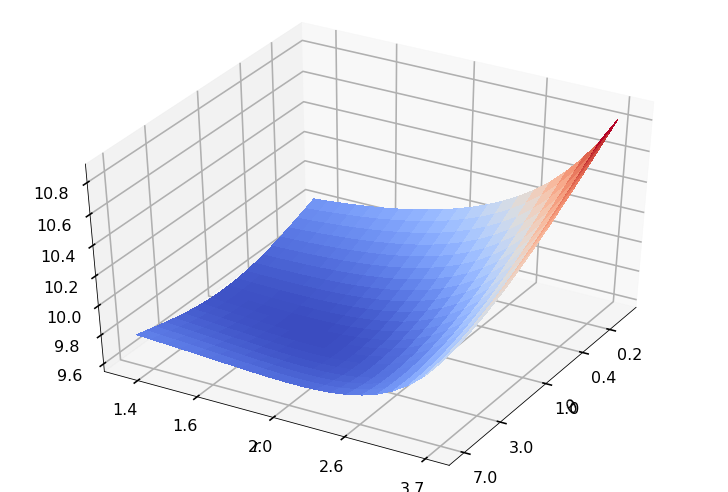
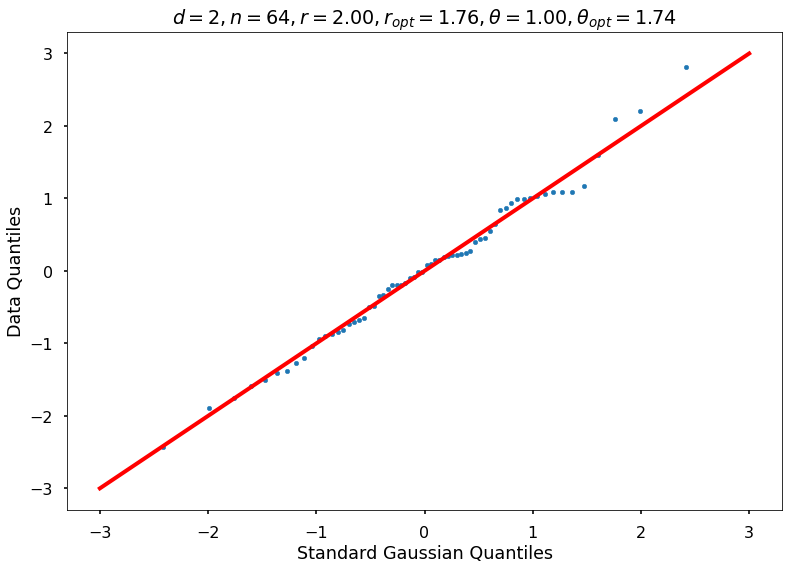
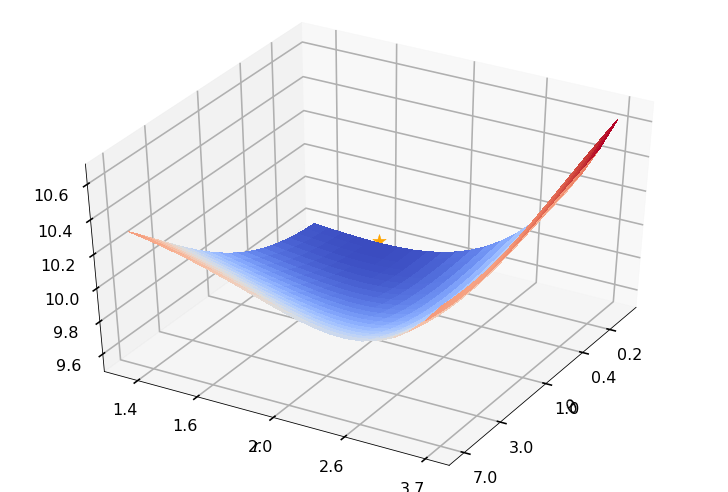
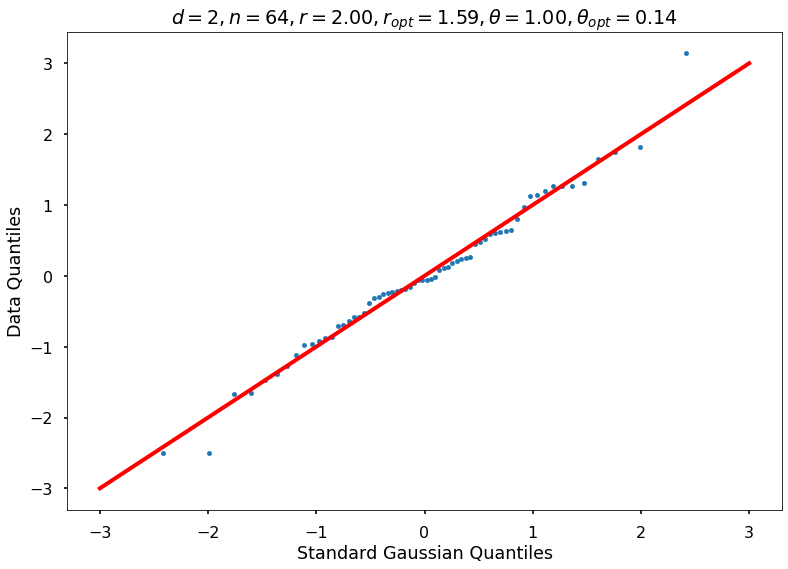
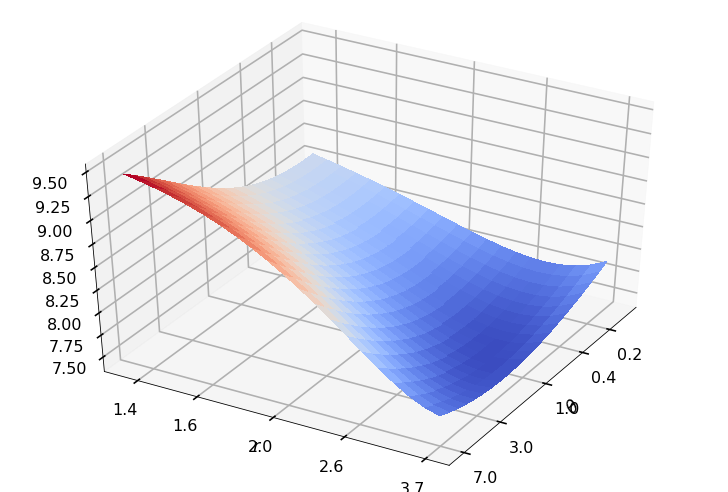
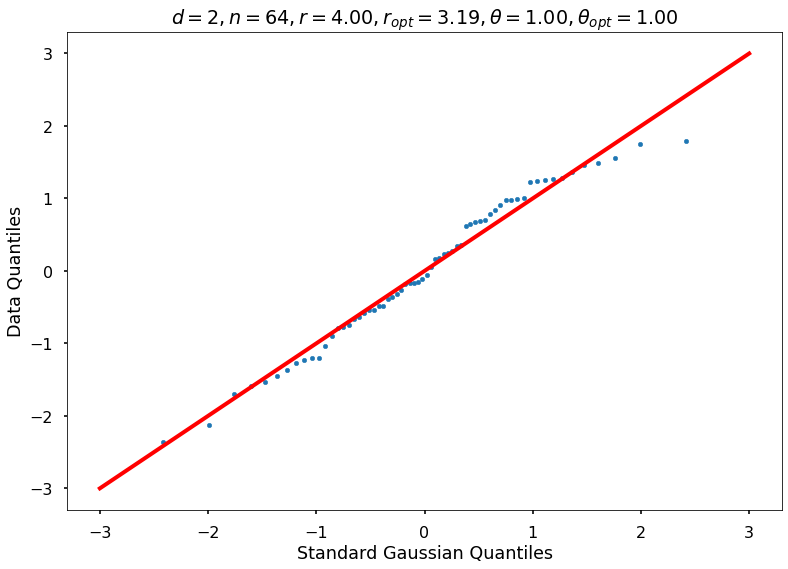
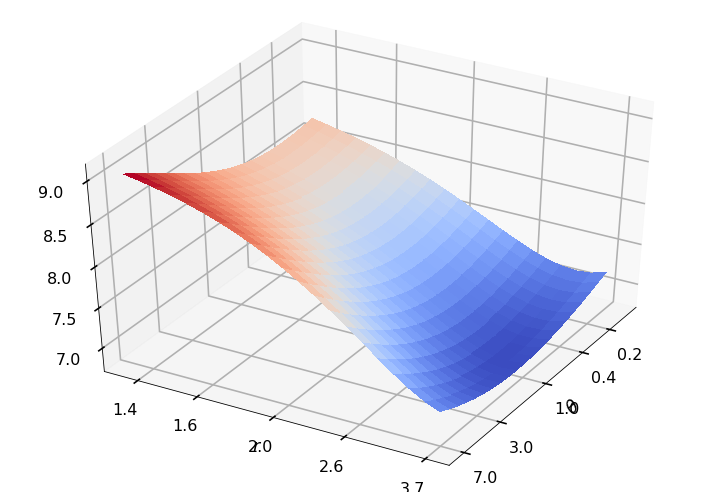
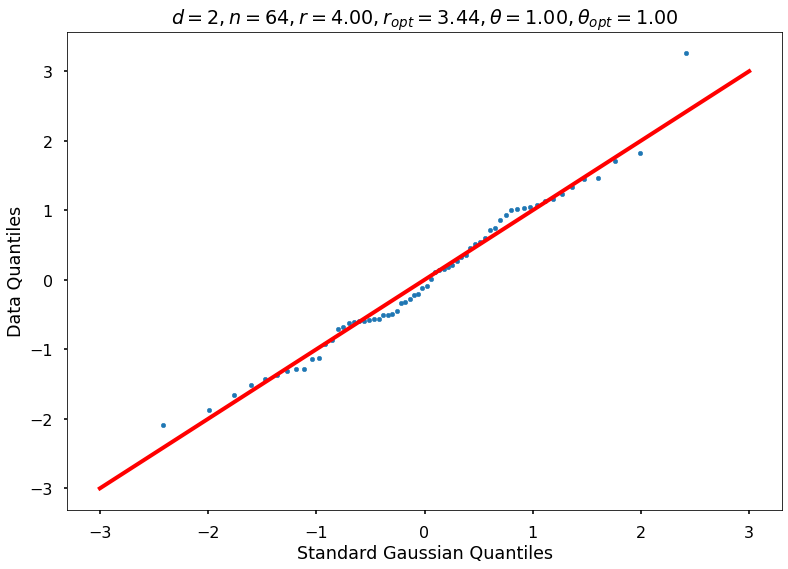
# close all the previous plots to freeup memory
plt.close('all')
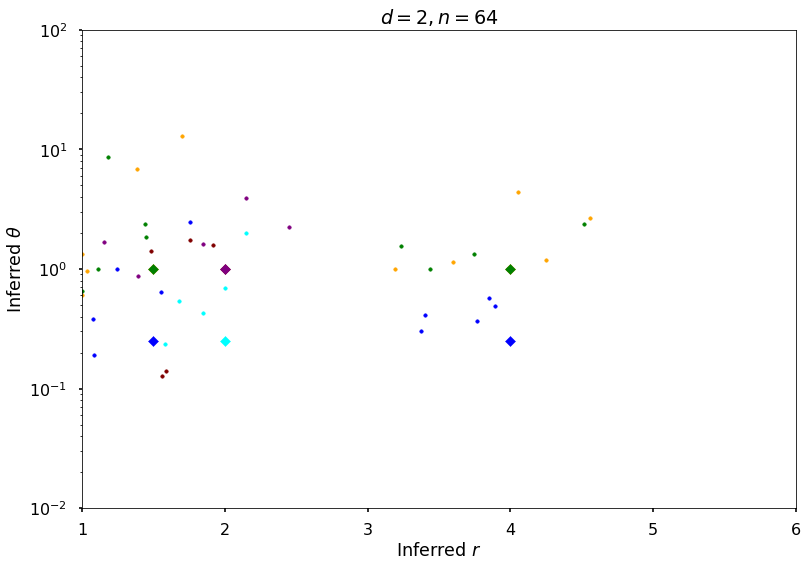
Plot additional figures for random function
figH, axH = plt.subplots()
colorArray = ['blue', 'orange', 'green', 'cyan', 'maroon', 'purple']
nColArray = len(colorArray)
for jjj in range(nrArr):
for kkk in range(nfPArr):
clrInd = np.mod(nfPArr * (jjj) + kkk, nColArray)
clr = colorArray[clrInd]
axH.scatter(rOptAll[jjj, kkk, :].reshape((nRep, 1)), thOptAll[jjj, kkk, :].reshape((nRep, 1)),
s=50, c=clr, marker='.')
axH.scatter(rArray[jjj], thetaAll[jjj, kkk], s=50, c=clr, marker='D')
axH.set(xlim=[1, 6], ylim=[0.01, 100])
axH.set_yscale('log')
axH.set_title(f'$d = {dim}, n = {npts}$')
axH.set_xlabel('Inferred $r$')
axH.set_ylabel('Inferred $\\theta$')
figH.savefig(f'{fName}-rthInfer-n-{npts}-d-{dim}.jpg')
# close all the previous plots to freeup memory
plt.close('all')
Example 3a: Keister integrand: npts = 64
## Keister example
fwh = 2
dim = 3
npts = 2 ** 6
nRep = 20
nPlot = 2
_, rOptAll, thOptAll, fName = gaussian_diagnostics_engine(fwh, dim, npts, None, None, nRep, nPlot)
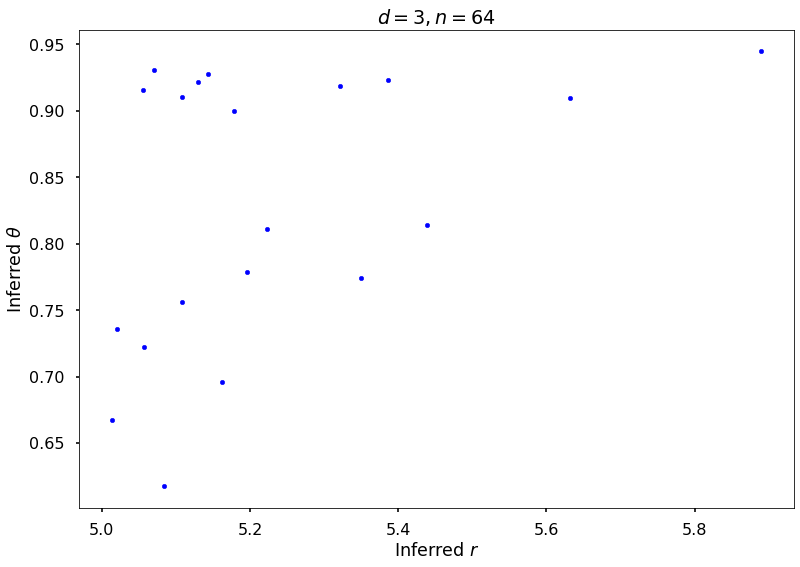
## Plot Keister example
figH = plt.figure()
plt.scatter(rOptAll, thOptAll, s=20, color='blue')
# axis([4 6 0.5 1.5])
# set(gca,'yscale','log')
plt.xlabel('Inferred $r$')
plt.ylabel('Inferred $\\theta$')
plt.title(f'$d = {dim}, n = {npts}$')
figH.savefig(f'{fName}-rthInfer-n-{npts}-d-{dim}.jpg')
10.918373367804714
10.74245342720698
r = None, rOpt = 5.17845, theta = None, thetaOpt = 0.90000
10.717466826946536
10.562731448359418
r = None, rOpt = 5.10797, theta = None, thetaOpt = 0.75631
10.688455595826237
10.539160528442594
r = None, rOpt = 5.02081, theta = None, thetaOpt = 0.73583
10.874548555226458
10.610842354541466
r = None, rOpt = 5.63220, theta = None, thetaOpt = 0.90923
10.740805174948838
10.557754768898818
r = None, rOpt = 5.19574, theta = None, thetaOpt = 0.77893
10.928140146153147
10.720109505682482
r = None, rOpt = 5.43883, theta = None, thetaOpt = 0.81368
10.936776084649589
10.748392943135848
r = None, rOpt = 5.32207, theta = None, thetaOpt = 0.91872
10.866161104206988
10.690655913547836
r = None, rOpt = 5.14400, theta = None, thetaOpt = 0.92759
10.613489865692868
10.45889197376518
r = None, rOpt = 5.08381, theta = None, thetaOpt = 0.61740
10.636181168972138
10.467815857368016
r = None, rOpt = 5.16286, theta = None, thetaOpt = 0.69577
10.728895335365538
10.585065856399083
r = None, rOpt = 5.05675, theta = None, thetaOpt = 0.72234
11.031579085996949
10.889786816011357
r = None, rOpt = 5.05532, theta = None, thetaOpt = 0.91526
10.866073731336623
10.548636876696989
r = None, rOpt = 5.88962, theta = None, thetaOpt = 0.94453
10.898763849532882
10.7428251348116
r = None, rOpt = 5.07111, theta = None, thetaOpt = 0.93066
11.04144749198928
10.892511325588508
r = None, rOpt = 5.10849, theta = None, thetaOpt = 0.91039
10.771539976900868
10.602263037779617
r = None, rOpt = 5.22262, theta = None, thetaOpt = 0.81070
10.919713159295926
10.703969564045602
r = None, rOpt = 5.38652, theta = None, thetaOpt = 0.92338
10.775915237177964
10.646594662551685
r = None, rOpt = 5.01369, theta = None, thetaOpt = 0.66747
11.02928162756276
10.874074219278276
r = None, rOpt = 5.12990, theta = None, thetaOpt = 0.92172
10.87752870870672
10.685153898271022
r = None, rOpt = 5.34982, theta = None, thetaOpt = 0.77387

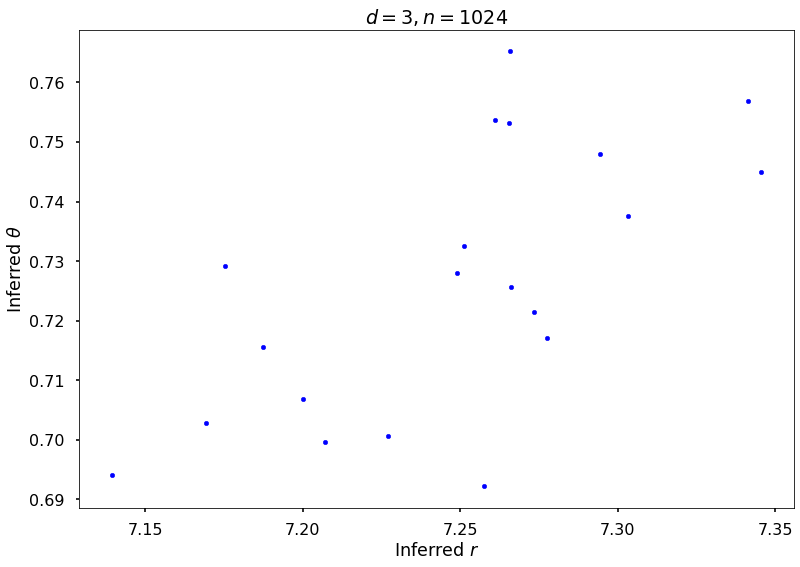
Example 3b: Keister integrand: npts = 1024
## Keister example
fwh = 2
dim = 3
npts = 2 ** 10
nRep = 20
nPlot = 2
_, rOptAll, thOptAll, fName = gaussian_diagnostics_engine(fwh, dim, npts, None, None, nRep, nPlot)
## Plot Keister example
figH = plt.figure()
plt.scatter(rOptAll, thOptAll, s=20, color='blue')
# axis([4 6 0.5 1.5])
# set(gca,'yscale','log')
plt.xlabel('Inferred $r$')
plt.ylabel('Inferred $\\theta$')
plt.title(f'$d = {dim}, n = {npts}$')
figH.savefig(f'{fName}-rthInfer-n-{npts}-d-{dim}.jpg')
10.596244143639485
7.142978946028139
r = None, rOpt = 7.18724, theta = None, thetaOpt = 0.71553
10.594856691086207
7.096409091118618
r = None, rOpt = 7.26586, theta = None, thetaOpt = 0.76520
10.591375134255042
7.0166323209172194
r = None, rOpt = 7.34554, theta = None, thetaOpt = 0.74504
10.595755571171377
7.067858016018031
r = None, rOpt = 7.34121, theta = None, thetaOpt = 0.75696
10.595982354998483
7.1228385587643395
r = None, rOpt = 7.22718, theta = None, thetaOpt = 0.70060
10.596298140666004
7.104717917027118
r = None, rOpt = 7.27766, theta = None, thetaOpt = 0.71703
10.592187612685716
7.0859774608781745
r = None, rOpt = 7.26092, theta = None, thetaOpt = 0.75369
10.596120527059021
7.10600296848717
r = None, rOpt = 7.25766, theta = None, thetaOpt = 0.69220
10.594256056513693
7.105374164104877
r = None, rOpt = 7.25124, theta = None, thetaOpt = 0.73247
10.596614398709978
7.137622143665308
r = None, rOpt = 7.20010, theta = None, thetaOpt = 0.70688
10.598230772383527
7.186852606944651
r = None, rOpt = 7.13933, theta = None, thetaOpt = 0.69409
10.597856894192905
7.126442092925792
r = None, rOpt = 7.27342, theta = None, thetaOpt = 0.72154
10.595526689101934
7.101590379268407
r = None, rOpt = 7.26605, theta = None, thetaOpt = 0.72566
10.593990233649055
7.128838039518072
r = None, rOpt = 7.17539, theta = None, thetaOpt = 0.72916
10.594285418816956
7.138947266052696
r = None, rOpt = 7.16931, theta = None, thetaOpt = 0.70287
10.597310034462971
7.117069509729076
r = None, rOpt = 7.30321, theta = None, thetaOpt = 0.73752
10.592677212556865
7.085904146817816
r = None, rOpt = 7.26545, theta = None, thetaOpt = 0.75314
10.592216478157543
7.05396738656259
r = None, rOpt = 7.29427, theta = None, thetaOpt = 0.74802
10.59392746212006
7.109982966182546
r = None, rOpt = 7.20702, theta = None, thetaOpt = 0.69972
10.595997284727325
7.111744447354244
r = None, rOpt = 7.24907, theta = None, thetaOpt = 0.72795