A Monte Carlo vs Quasi-Monte Carlo Comparison
Monte Carlo algorithms work on independent identically distributed (IID) points while Quasi-Monte Carlo algorithms work on low discrepancy (LD) sequences. LD generators, such as those for the lattice and Sobol’ sequences, provide samples whose space filling properties can be exploited by Quasi-Monte Carlo algorithms.
import pandas as pd
pd.options.display.float_format = '{:.2e}'.format
from matplotlib import pyplot as plt
import matplotlib
%matplotlib inline
plt.rc('font', size=16) # controls default text sizes
plt.rc('axes', titlesize=16) # fontsize of the axes title
plt.rc('axes', labelsize=16) # fontsize of the x and y labels
plt.rc('xtick', labelsize=16) # fontsize of the tick labels
plt.rc('ytick', labelsize=16) # fontsize of the tick labels
plt.rc('legend', fontsize=16) # legend fontsize
plt.rc('figure', titlesize=16) # fontsize of the figure title
Vary Absolute Tolerance
Testing Parameters - relative tolerance = 0 - Results averaged over 3 trials
Keister Integrand - 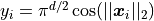 -
- 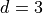
Gaussian True Measure -
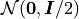
df = pd.read_csv('../workouts/mc_vs_qmc/out/vary_abs_tol.csv')
df['Problem'] = df['Stopping Criterion'] + ' ' + df['Distribution'] + ' (' + df['MC/QMC'] + ')'
df = df.drop(['Stopping Criterion','Distribution','MC/QMC'],axis=1)
problems = ['CubMCCLT IIDStdUniform (MC)',
'CubMCG IIDStdUniform (MC)',
'CubQMCCLT Sobol (QMC)',
'CubQMCLatticeG Lattice (QMC)',
'CubQMCSobolG Sobol (QMC)']
df = df[df['Problem'].isin(problems)]
df['abs_tol'] = df['abs_tol'].round(4)
df_grouped = df.groupby(['Problem'])
df_abs_tols = df_grouped['abs_tol'].apply(list).reset_index(name='abs_tol')
df_samples = df_grouped['n_samples'].apply(list).reset_index(name='n')
df_times = df.groupby(['Problem'])['time'].apply(list).reset_index(name='time')
df[df['abs_tol'].isin([.01,.05,.1])].set_index('Problem')
| abs_tol | solution | n_samples | time | |
|---|---|---|---|---|
| Problem | ||||
| CubMCCLT IIDStdUniform (MC) | 1.00e-02 | 2.17e+00 | 4.90e+05 | 9.05e-02 |
| CubMCCLT IIDStdUniform (MC) | 5.00e-02 | 2.14e+00 | 2.16e+04 | 5.49e-03 |
| CubMCCLT IIDStdUniform (MC) | 1.00e-01 | 2.15e+00 | 6.93e+03 | 2.50e-03 |
| CubMCG IIDStdUniform (MC) | 1.00e-02 | 2.17e+00 | 6.27e+05 | 1.11e-01 |
| CubMCG IIDStdUniform (MC) | 5.00e-02 | 2.14e+00 | 4.09e+04 | 8.46e-03 |
| CubMCG IIDStdUniform (MC) | 1.00e-01 | 2.14e+00 | 1.48e+04 | 5.97e-03 |
| CubQMCCLT Sobol (QMC) | 1.00e-02 | 2.17e+00 | 4.10e+03 | 3.89e-03 |
| CubQMCCLT Sobol (QMC) | 5.00e-02 | 2.17e+00 | 4.10e+03 | 3.14e-03 |
| CubQMCCLT Sobol (QMC) | 1.00e-01 | 2.17e+00 | 4.10e+03 | 3.11e-03 |
| CubQMCLatticeG Lattice (QMC) | 1.00e-02 | 2.17e+00 | 2.05e+03 | 5.67e-03 |
| CubQMCLatticeG Lattice (QMC) | 5.00e-02 | 2.17e+00 | 1.02e+03 | 3.80e-03 |
| CubQMCLatticeG Lattice (QMC) | 1.00e-01 | 2.17e+00 | 1.02e+03 | 3.72e-03 |
| CubQMCSobolG Sobol (QMC) | 1.00e-02 | 2.17e+00 | 2.05e+03 | 2.01e-03 |
| CubQMCSobolG Sobol (QMC) | 5.00e-02 | 2.17e+00 | 1.02e+03 | 1.05e-03 |
| CubQMCSobolG Sobol (QMC) | 1.00e-01 | 2.17e+00 | 1.02e+03 | 1.05e-03 |
fig,ax = plt.subplots(nrows=1, ncols=2, figsize=(18, 5))
for problem in problems:
abs_tols = df_abs_tols[df_abs_tols['Problem']==problem]['abs_tol'].tolist()[0]
samples = df_samples[df_samples['Problem']==problem]['n'].tolist()[0]
times = df_times[df_times['Problem']==problem]['time'].tolist()[0]
ax[0].plot(abs_tols,samples,label=problem)
ax[1].plot(abs_tols,times,label=problem)
for ax_i in ax:
ax_i.set_xscale('log', base=10)
ax_i.set_yscale('log', base=10)
ax_i.spines['right'].set_visible(False)
ax_i.spines['top'].set_visible(False)
ax_i.set_xlabel('Absolute Tolerance')
ax[0].legend(loc='upper right', frameon=False,ncol=1,bbox_to_anchor=(2.8,1))
ax[0].set_ylabel('Total Samples')
ax[1].set_ylabel('Runtime')
fig.suptitle('Comparing Absolute Tolerances')
plt.subplots_adjust(wspace=.15, hspace=0);
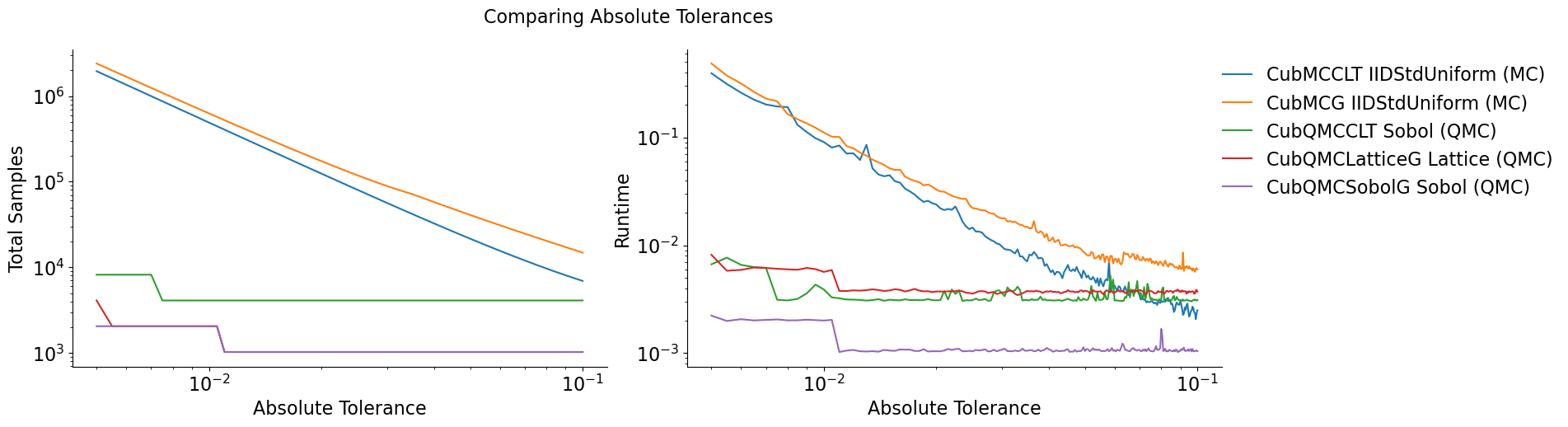
Quasi-Monte Carlo takes less time and fewer samples to achieve the same
accuracy as regular Monte Carlo The number of points for Monte Carlo
algorithms is 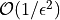 while quasi-Monte Carlo
algorithms can be as efficient as
while quasi-Monte Carlo
algorithms can be as efficient as 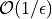
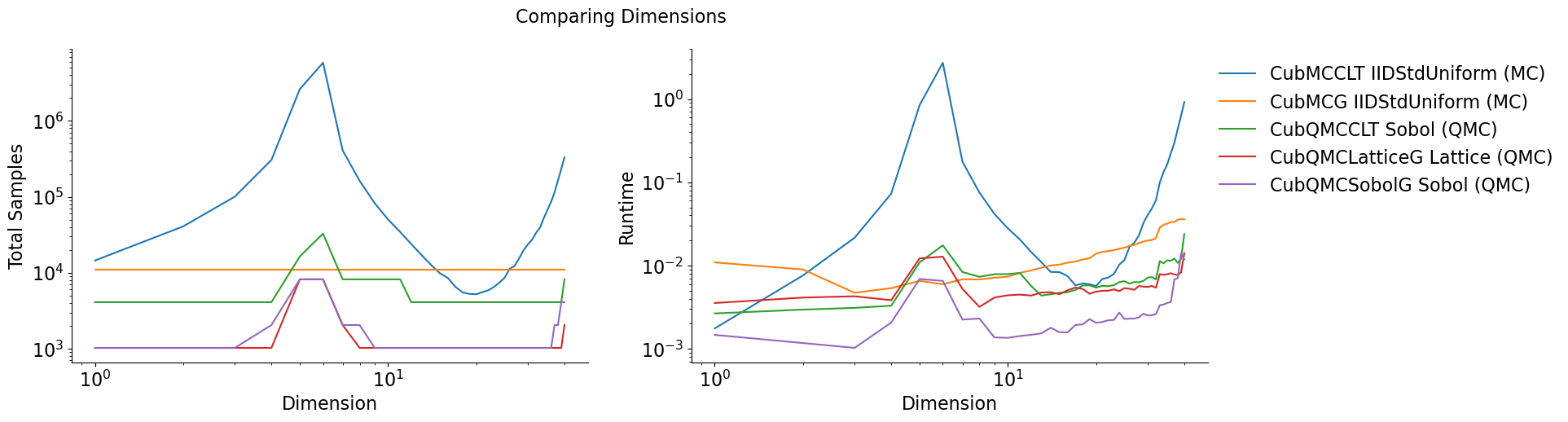
Vary Dimension
Testing Parameters - absolute tolerance = 0 - relative tolerance = .01 - Results averaged over 3 trials
Keister Integrand - 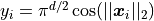
Gaussian True Measure -
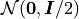
df = pd.read_csv('../workouts/mc_vs_qmc/out/vary_dimension.csv')
df['Problem'] = df['Stopping Criterion'] + ' ' + df['Distribution'] + ' (' + df['MC/QMC'] + ')'
df = df.drop(['Stopping Criterion','Distribution','MC/QMC'],axis=1)
problems = ['CubMCCLT IIDStdUniform (MC)',
'CubMCG IIDStdUniform (MC)',
'CubQMCCLT Sobol (QMC)',
'CubQMCLatticeG Lattice (QMC)',
'CubQMCSobolG Sobol (QMC)']
df = df[df['Problem'].isin(problems)]
df_grouped = df.groupby(['Problem'])
df_dims = df_grouped['dimension'].apply(list).reset_index(name='dimension')
df_samples = df_grouped['n_samples'].apply(list).reset_index(name='n')
df_times = df.groupby(['Problem'])['time'].apply(list).reset_index(name='time')
df[df['dimension'].isin([10,30])].set_index('Problem')
| dimension | solution | n_samples | time | |
|---|---|---|---|---|
| Problem | ||||
| CubMCCLT IIDStdUniform (MC) | 10 | -1.55e+02 | 5.04e+04 | 2.80e-02 |
| CubMCCLT IIDStdUniform (MC) | 30 | -1.94e+07 | 2.37e+04 | 4.05e-02 |
| CubMCG IIDStdUniform (MC) | 10 | -1.56e+02 | 1.10e+04 | 7.36e-03 |
| CubMCG IIDStdUniform (MC) | 30 | -1.94e+07 | 1.10e+04 | 2.00e-02 |
| CubQMCCLT Sobol (QMC) | 10 | -1.54e+02 | 8.19e+03 | 7.91e-03 |
| CubQMCCLT Sobol (QMC) | 30 | -1.94e+07 | 4.10e+03 | 7.15e-03 |
| CubQMCLatticeG Lattice (QMC) | 10 | -1.55e+02 | 1.02e+03 | 4.40e-03 |
| CubQMCLatticeG Lattice (QMC) | 30 | -1.94e+07 | 1.02e+03 | 5.54e-03 |
| CubQMCSobolG Sobol (QMC) | 10 | -1.54e+02 | 1.02e+03 | 1.36e-03 |
| CubQMCSobolG Sobol (QMC) | 30 | -1.94e+07 | 1.02e+03 | 2.51e-03 |
fig,ax = plt.subplots(nrows=1, ncols=2, figsize=(18, 5))
for problem in problems:
dimension = df_dims[df_dims['Problem']==problem]['dimension'].tolist()[0]
samples = df_samples[df_samples['Problem']==problem]['n'].tolist()[0]
times = df_times[df_times['Problem']==problem]['time'].tolist()[0]
ax[0].plot(dimension,samples,label=problem)
ax[1].plot(dimension,times,label=problem)
for ax_i in ax:
ax_i.set_xscale('log', base=10)
ax_i.set_yscale('log', base=10)
ax_i.spines['right'].set_visible(False)
ax_i.spines['top'].set_visible(False)
ax_i.set_xlabel('Dimension')
ax[0].legend(loc='upper right', frameon=False,ncol=1,bbox_to_anchor=(2.9,1))
ax[0].set_ylabel('Total Samples')
ax[1].set_ylabel('Runtime')
fig.suptitle('Comparing Dimensions');